AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in arabidopsis
- PMID: 24424095
- PMCID: PMC3963575
- DOI: 10.1105/tpc.113.120048
AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in arabidopsis
Abstract
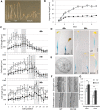
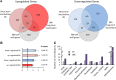
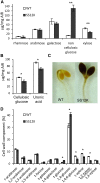
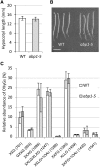
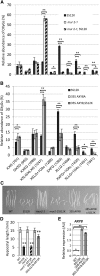
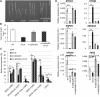
Cell expansion is an increase in cell size and thus plays an essential role in plant growth and development. Phytohormones and the primary plant cell wall play major roles in the complex process of cell expansion. In shoot tissues, cell expansion requires the auxin receptor AUXIN BINDING PROTEIN1 (ABP1), but the mechanism by which ABP1 affects expansion remains unknown. We analyzed the effect of functional inactivation of ABP1 on transcriptomic changes in dark-grown hypocotyls and investigated the consequences of gene expression on cell wall composition and cell expansion. Molecular and genetic evidence indicates that ABP1 affects the expression of a broad range of cell wall-related genes, especially cell wall remodeling genes, mainly via an SCF(TIR/AFB)-dependent pathway. ABP1 also functions in the modulation of hemicellulose xyloglucan structure. Furthermore, fucosidase-mediated defucosylation of xyloglucan, but not biosynthesis of nonfucosylated xyloglucan, rescued dark-grown hypocotyl lengthening of ABP1 knockdown seedlings. In muro remodeling of xyloglucan side chains via an ABP1-dependent pathway appears to be of critical importance for temporal and spatial control of cell expansion.
Figures

References
-
- Augur C., Benhamou N., Darvill A., Albersheim P. (1993). Purification, characterization, and cell wall localization of an alpha-fucosidase that inactivates a xyloglucan oligosaccharin. Plant J. 3: 415–426 - PubMed
-
- Barbier-Brygoo H., Zimmermann S., Thomine S., White I.R., Millner P., Guern J. (1996). Elementary auxin response chains at the plasma membrane involve external abp1 and multiple electrogenic ion transport proteins. Plant Growth Regul. 18: 23–28
-
- Blumenkrantz N., Asboe-Hansen G. (1973). New method for quantitative determination of uronic acids. Anal. Biochem. 54: 484–489 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

