A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth
- PMID: 24424321
- PMCID: PMC3938631
- DOI: 10.1104/pp.113.230896
A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth
Abstract
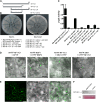
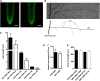
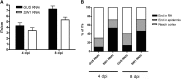
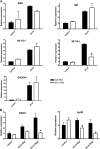
A C subunit of the heterotrimeric nuclear factor Y (NF-YC1) was shown to play a key role in nodule organogenesis and bacterial infection during the nitrogen fixing symbiosis established between common bean (Phaseolus vulgaris) and Rhizobium etli. To identify other proteins involved in this process, we used the yeast (Saccharomyces cerevisiae) two-hybrid system to screen for NF-YC1-interacting proteins. One of the positive clones encodes a member of the Phytochrome A Signal Transduction1 subfamily of GRAS (for Gibberellic Acid-Insensitive (GAI), Repressor of GAI, and Scarecrow) transcription factors. The protein, named Scarecrow-like13 Involved in Nodulation (SIN1), localizes both to the nucleus and the cytoplasm, but in transgenic Nicotiana benthamiana cells, bimolecular fluorescence complementation suggested that the interaction with NF-YC1 takes place predominantly in the nucleus. SIN1 is expressed in aerial and root tissues, with higher levels in roots and nodules. Posttranscriptional gene silencing of SIN1 using RNA interference (RNAi) showed that the product of this gene is involved in lateral root elongation. However, root cell organization, density of lateral roots, and the length of root hairs were not affected by SIN1 RNAi. In addition, the expression of the RNAi of SIN1 led to a marked reduction in the number and size of nodules formed upon inoculation with R. etli and affected the progression of infection threads toward the nodule primordia. Expression of NF-YA1 and the G2/M transition cell cycle genes Cyclin B and Cell Division Cycle2 was reduced in SIN1 RNAi roots. These data suggest that SIN1 plays a role in lateral root elongation and the establishment of root symbiosis in common bean.
Figures



References
-
- Bolle C. (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218: 683–692 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

