Dom34-Hbs1 mediated dissociation of inactive 80S ribosomes promotes restart of translation after stress
- PMID: 24424461
- PMCID: PMC3989619
- DOI: 10.1002/embj.201386123
Dom34-Hbs1 mediated dissociation of inactive 80S ribosomes promotes restart of translation after stress
Abstract
Following translation termination, ribosomal subunits dissociate to become available for subsequent rounds of protein synthesis. In many translation-inhibiting stress conditions, e.g. glucose starvation in yeast, free ribosomal subunits reassociate to form a large pool of non-translating 80S ribosomes stabilized by the 'clamping' Stm1 factor. The subunits of these inactive ribosomes need to be mobilized for translation restart upon stress relief. The Dom34-Hbs1 complex, together with the Rli1 NTPase (also known as ABCE1), have been shown to split ribosomes stuck on mRNAs in the context of RNA quality control mechanisms. Here, using in vitro and in vivo methods, we report a new role for the Dom34-Hbs1 complex and Rli1 in dissociating inactive ribosomes, thereby facilitating translation restart in yeast recovering from glucose starvation stress. Interestingly, we found that this new role is not restricted to stress conditions, indicating that in growing yeast there is a dynamic pool of inactive ribosomes that needs to be split by Dom34-Hbs1 and Rli1 to participate in protein synthesis. We propose that this provides a new level of translation regulation.
Figures
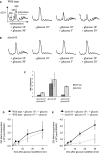
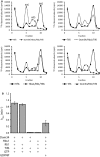
Dom34-Hbs1 and Rli1 dissociate ribosomes from glucose-starved yeast in vitro. 32P-labeled 80S ribosomes purified from glucose-starved yeast were incubated with the indicated proteins in the presence of ATP and GTP or GDPNP. After 15 min of incubation dissociation was monitored by sucrose density gradient centrifugation and scintillation counting of collected fractions.
Observed rate constants were determined by monitoring the fraction of dissociated ribosomes in (A) over time on a native gel system (see Supplementary Fig S2). Rate constants were normalized for endpoints. Means ± standard deviations of three independent experiments are shown.



Similar articles
-
Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3' end of aberrant mRNA.Mol Cell. 2012 May 25;46(4):518-29. doi: 10.1016/j.molcel.2012.03.013. Epub 2012 Apr 11. Mol Cell. 2012. PMID: 22503425
-
Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay.Science. 2010 Oct 15;330(6002):369-72. doi: 10.1126/science.1192430. Science. 2010. PMID: 20947765 Free PMC article.
-
Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution.Nat Commun. 2016 Dec 20;7:13521. doi: 10.1038/ncomms13521. Nat Commun. 2016. PMID: 27995908 Free PMC article.
-
No-go decay: a quality control mechanism for RNA in translation.Wiley Interdiscip Rev RNA. 2010 Jul-Aug;1(1):132-41. doi: 10.1002/wrna.17. Epub 2010 May 6. Wiley Interdiscip Rev RNA. 2010. PMID: 21956910 Review.
-
The elongation, termination, and recycling phases of translation in eukaryotes.Cold Spring Harb Perspect Biol. 2012 Jul 1;4(7):a013706. doi: 10.1101/cshperspect.a013706. Cold Spring Harb Perspect Biol. 2012. PMID: 22751155 Free PMC article. Review.
Cited by
-
Ribosome recycling is coordinated by processive events in two asymmetric ATP sites of ABCE1.Life Sci Alliance. 2018 Jun 14;1(3):e201800095. doi: 10.26508/lsa.201800095. Life Sci Alliance. 2018. PMID: 30198020 Free PMC article.
-
Regulation of eukaryotic translation initiation factor 6 dynamics through multisite phosphorylation by GSK3.J Biol Chem. 2020 Sep 4;295(36):12796-12813. doi: 10.1074/jbc.RA120.013324. Epub 2020 Jul 23. J Biol Chem. 2020. PMID: 32703900 Free PMC article.
-
Efficient analysis of mammalian polysomes in cells and tissues using Ribo Mega-SEC.Elife. 2018 Aug 10;7:e36530. doi: 10.7554/eLife.36530. Elife. 2018. PMID: 30095066 Free PMC article.
-
Structures of translationally inactive mammalian ribosomes.Elife. 2018 Oct 24;7:e40486. doi: 10.7554/eLife.40486. Elife. 2018. PMID: 30355441 Free PMC article.
-
Decoupling Yeast Cell Division and Stress Defense Implicates mRNA Repression in Translational Reallocation during Stress.Curr Biol. 2018 Aug 20;28(16):2673-2680.e4. doi: 10.1016/j.cub.2018.06.044. Epub 2018 Aug 2. Curr Biol. 2018. PMID: 30078561 Free PMC article.
References
-
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. - PubMed
-
- Andersen DS, Leevers SJ. The essential Drosophila ATP-binding cassette domain protein, pixie, binds the 40 S ribosome in an ATP-dependent manner and is required for translation initiation. J Biol Chem. 2007;282:14752–14760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

