A high-definition view of functional genetic variation from natural yeast genomes
- PMID: 24425782
- PMCID: PMC3969562
- DOI: 10.1093/molbev/msu037
A high-definition view of functional genetic variation from natural yeast genomes
Abstract
The question of how genetic variation in a population influences phenotypic variation and evolution is of major importance in modern biology. Yet much is still unknown about the relative functional importance of different forms of genome variation and how they are shaped by evolutionary processes. Here we address these questions by population level sequencing of 42 strains from the budding yeast Saccharomyces cerevisiae and its closest relative S. paradoxus. We find that genome content variation, in the form of presence or absence as well as copy number of genetic material, is higher within S. cerevisiae than within S. paradoxus, despite genetic distances as measured in single-nucleotide polymorphisms being vastly smaller within the former species. This genome content variation, as well as loss-of-function variation in the form of premature stop codons and frameshifting indels, is heavily enriched in the subtelomeres, strongly reinforcing the relevance of these regions to functional evolution. Genes affected by these likely functional forms of variation are enriched for functions mediating interaction with the external environment (sugar transport and metabolism, flocculation, metal transport, and metabolism). Our results and analyses provide a comprehensive view of genomic diversity in budding yeast and expose surprising and pronounced differences between the variation within S. cerevisiae and that within S. paradoxus. We also believe that the sequence data and de novo assemblies will constitute a useful resource for further evolutionary and population genomics studies.
Keywords: functional variation; genome evolution; loss-of-function variants; population genomics; subtelomeres; yeast.
Figures
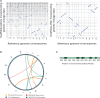
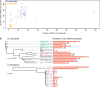



Comment in
-
New sequencing tools give a close look at yeast evolution.Mol Biol Evol. 2014 Apr;31(4):1056-7. doi: 10.1093/molbev/msu060. Epub 2014 Feb 17. Mol Biol Evol. 2014. PMID: 24536042 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

