Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection
- PMID: 24425785
- PMCID: PMC3989421
- DOI: 10.1158/1541-7786.MCR-13-0396
Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection
Abstract
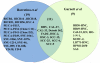
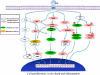
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common type of cancer worldwide. The increasing amount of genomic information on human tumors and cell lines provides more biologic data to design preclinical studies. We and others previously reported whole-exome sequencing data of 106 HNSCC primary tumors. In 2012, high-throughput genomic data and pharmacologic profiling of anticancer drugs of hundreds of cancer cell lines were reported. Here, we compared the genomic data of 39 HNSCC cell lines with the genomic findings in 106 HNSCC tumors. Amplification of eight genes (PIK3CA, EGFR, CCND2, KDM5A, ERBB2, PMS1, FGFR1, and WHSCIL1) and deletion of five genes (CDKN2A, SMAD4, NOTCH2, NRAS, and TRIM33) were found in both HNSCC cell lines and tumors. Seventeen genes were only mutated in HNSCC cell lines (>10%), suggesting that these mutations may arise through immortalization in tissue culture. Conversely, 11 genes were only mutated in >10% of human HNSCC tumors. Several mutant genes in the EGF receptor (EGFR) pathway are shared both in cell lines and in tumors. Pharmacologic profiling of eight anticancer agents in six HNSCC cell lines suggested that PIK3CA mutation may serve as a predictive biomarker for the drugs targeting the EGFR/PI3K pathway. These findings suggest that a correlation of gene mutations between HNSCC cell lines and human tumors may be used to guide the selection of preclinical models for translational research.
Implications: These findings suggest that a correlation of gene mutations between HNSCC cell lines and human tumors may be used to guide the selection of preclinical models for translational research.
Mol Cancer Res; 12(4); 571-82. ©2014 AACR.
Figures






References
-
- René Leemans C, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011 Jan;11(1):9–22. - PubMed
-
- Kutler DI, Auerbach AD, Satagopan J, Giampietro PF, Batish SD, Huvos AG, et al. High incidence of head and neck squamous cell carcinoma in patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg. 2003 Jan;129(1):106–12. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

