CD2-associated protein (CD2AP) enhances casitas B lineage lymphoma-3/c (Cbl-3/c)-mediated Ret isoform-specific ubiquitination and degradation via its amino-terminal Src homology 3 domains
- PMID: 24425877
- PMCID: PMC3953248
- DOI: 10.1074/jbc.M113.537878
CD2-associated protein (CD2AP) enhances casitas B lineage lymphoma-3/c (Cbl-3/c)-mediated Ret isoform-specific ubiquitination and degradation via its amino-terminal Src homology 3 domains
Abstract
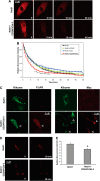
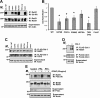
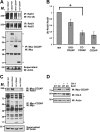
Ret is the receptor tyrosine kinase for the glial cell line-derived neurotrophic factor (GDNF) family of neuronal growth factors. Upon activation by GDNF, Ret is rapidly polyubiquitinated and degraded. This degradation process is isoform-selective, with the longer Ret51 isoform exhibiting different degradation kinetics than the shorter isoform, Ret9. In sympathetic neurons, Ret degradation is induced, at least in part, by a complex consisting of the adaptor protein CD2AP and the E3-ligase Cbl-3/c. Knockdown of Cbl-3/c using siRNA reduced the GDNF-induced ubiquitination and degradation of Ret51 in neurons and podocytes, suggesting that Cbl-3/c was a predominant E3 ligase for Ret. Coexpression of CD2AP with Cbl-3/c augmented the ubiquitination of Ret51 as compared with the expression of Cbl-3/c alone. Ret51 ubiquitination by the CD2AP·Cbl-3/c complex required a functional ring finger and TKB domain in Cbl-3/c. The SH3 domains of CD2AP were sufficient to drive the Cbl-3/c-dependent ubiquitination of Ret51, whereas the carboxyl-terminal coiled-coil domain of CD2AP was dispensable. Interestingly, activated Ret induced the degradation of CD2AP, but not Cbl-3/c, suggesting a potential inhibitory feedback mechanism. There were only two major ubiquitination sites in Ret51, Lys(1060) and Lys(1107), and the combined mutation of these lysines almost completely eliminated both the ubiquitination and degradation of Ret51. Ret9 was not ubiquitinated by the CD2AP·Cbl-3/c complex, suggesting that Ret9 was down-regulated by a fundamentally different mechanism. Taken together, these results suggest that only the SH3 domains of CD2AP were necessary to enhance the E3 ligase activity of Cbl-3/c toward Ret51.
Keywords: Neurodevelopment; Neurotrophic Factor; Protein Degradation; Receptor Tyrosine Kinase; Ubiquitination.
Figures



Similar articles
-
CD2AP and Cbl-3/Cbl-c constitute a critical checkpoint in the regulation of ret signal transduction.J Neurosci. 2008 Aug 27;28(35):8789-800. doi: 10.1523/JNEUROSCI.2738-08.2008. J Neurosci. 2008. PMID: 18753381 Free PMC article.
-
Distinct turnover of alternatively spliced isoforms of the RET kinase receptor mediated by differential recruitment of the Cbl ubiquitin ligase.J Biol Chem. 2005 Apr 8;280(14):13442-9. doi: 10.1074/jbc.M500507200. Epub 2005 Jan 27. J Biol Chem. 2005. PMID: 15677445
-
Differential recruitment of E3 ubiquitin ligase complexes regulates RET isoform internalization.J Cell Sci. 2017 Oct 1;130(19):3282-3296. doi: 10.1242/jcs.203885. Epub 2017 Aug 9. J Cell Sci. 2017. PMID: 28794017
-
Ubiquitin ligase Cbl-b and inhibitory Cblin peptides.Biochim Biophys Acta Proteins Proteom. 2020 Nov;1868(11):140495. doi: 10.1016/j.bbapap.2020.140495. Epub 2020 Jul 12. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 32663526 Review.
-
E3 ubiquitin ligase Cbl-b in innate and adaptive immunity.Cell Cycle. 2014;13(12):1875-84. doi: 10.4161/cc.29213. Epub 2014 May 14. Cell Cycle. 2014. PMID: 24875217 Free PMC article. Review.
Cited by
-
ATF4 destabilizes RET through nonclassical GRP78 inhibition to enhance chemosensitivity to bortezomib in human osteosarcoma.Theranostics. 2019 Aug 14;9(21):6334-6353. doi: 10.7150/thno.36818. eCollection 2019. Theranostics. 2019. PMID: 31534554 Free PMC article.
-
Noncanonical function of folate through folate receptor 1 during neural tube formation.Nat Commun. 2024 Feb 22;15(1):1642. doi: 10.1038/s41467-024-45775-1. Nat Commun. 2024. PMID: 38388461 Free PMC article.
-
Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).J Biol Chem. 2015 Oct 16;290(42):25275-92. doi: 10.1074/jbc.M115.637207. Epub 2015 Aug 20. J Biol Chem. 2015. PMID: 26296892 Free PMC article.
-
Non-canonical Ret signaling augments p75-mediated cell death in developing sympathetic neurons.J Cell Biol. 2018 Sep 3;217(9):3237-3253. doi: 10.1083/jcb.201703120. Epub 2018 Jul 17. J Cell Biol. 2018. PMID: 30018091 Free PMC article.
-
CD2AP at the junction of nephropathy and Alzheimer's disease.Mol Neurodegener. 2025 Jun 4;20(1):63. doi: 10.1186/s13024-025-00852-x. Mol Neurodegener. 2025. PMID: 40462155 Free PMC article. Review.
References
-
- Airaksinen M. S., Saarma M. (2002) The GDNF family. Signalling, biological functions and therapeutic value. Nat. Rev. Neurosci. 3, 383–394 - PubMed
-
- Baloh R. H., Enomoto H., Johnson E. M., Jr., Milbrandt J. (2000) The GDNF family ligands and receptors-implications for neural development. Curr. Opin. Neurobiol. 10, 103–110 - PubMed
-
- Bespalov M. M., Saarma M. (2007) GDNF family receptor complexes are emerging drug targets. Trends Pharmacol. Sci. 28, 68–74 - PubMed
-
- Wells S. A., Jr., Santoro M. (2009) Targeting the RET pathway in thyroid cancer. Clin. Cancer Res. 15, 7119–7123 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

