Next-generation sequencing of disseminated tumor cells
- PMID: 24427740
- PMCID: PMC3876274
- DOI: 10.3389/fonc.2013.00320
Next-generation sequencing of disseminated tumor cells
Abstract
Disseminated tumor cells (DTCs) detected in the bone marrow have been shown as an independent prognostic factor for women with breast cancer. However, the mechanisms behind the tumor cell dissemination are still unclear and more detailed knowledge is needed to fully understand why some cells remain dormant and others metastasize. Sequencing of single cells has opened for the possibility to dissect the genetic content of subclones of a primary tumor, as well as DTCs. Previous studies of genetic changes in DTCs have employed single-cell array comparative genomic hybridization which provides information about larger aberrations. To date, next-generation sequencing provides the possibility to discover new, smaller, and copy neutral genetic changes. In this study, we performed whole-genome amplification and subsequently next-generation sequencing to analyze DTCs from two breast cancer patients. We compared copy-number profiles of the DTCs and the corresponding primary tumor generated from sequencing and SNP-comparative genomic hybridization (CGH) data, respectively. While one tumor revealed mostly whole-arm gains and losses, the other had more complex alterations, as well as subclonal amplification and deletions. Whole-arm gains or losses in the primary tumor were in general also observed in the corresponding DTC. Both primary tumors showed amplification of chromosome 1q and deletion of parts of chromosome 16q, which was recaptured in the corresponding DTCs. Interestingly, clear differences were also observed, indicating that the DTC underwent further evolution at the copy-number level. This study provides a proof-of-principle for sequencing of DTCs and correlation with primary copy-number profiles. The analyses allow insight into tumor cell dissemination and show ongoing copy-number evolution in DTCs compared to the primary tumors.
Keywords: circulating tumor cells; clonal evolution; disseminating tumor cells; single tumor cell sequencing; tumor heterogeneity.
Figures
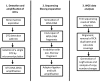
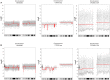
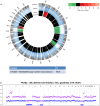


References
-
- Braun S, Kentenich C, Janni W, Hepp F, de Waal J, Willgeroth F, et al. Lack of effect of adjuvant chemotherapy on the elimination of single dormant tumor cells in bone marrow of high-risk breast cancer patients. J Clin Oncol (2000) 18(1):80–6 - PubMed
-
- Xenidis N, Perraki M, Kafousi M, Apostolaki S, Bolonaki I, Stathopoulou A, et al. Predictive and prognostic value of peripheral blood cytokeratin-19 mRNA-positive cells detected by real-time polymerase chain reaction in node-negative breast cancer patients. J Clin Oncol (2006) 24(23):3756–6210.1200/JCO.2005.04.5948 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

