Microscale sulfur cycling in the phototrophic pink berry consortia of the Sippewissett Salt Marsh
- PMID: 24428801
- PMCID: PMC4262008
- DOI: 10.1111/1462-2920.12388
Microscale sulfur cycling in the phototrophic pink berry consortia of the Sippewissett Salt Marsh
Abstract
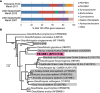
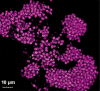
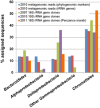
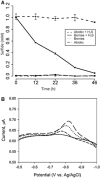
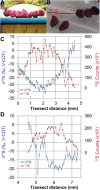
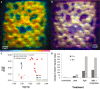
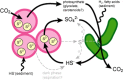
Microbial metabolism is the engine that drives global biogeochemical cycles, yet many key transformations are carried out by microbial consortia over short spatiotemporal scales that elude detection by traditional analytical approaches. We investigate syntrophic sulfur cycling in the 'pink berry' consortia of the Sippewissett Salt Marsh through an integrative study at the microbial scale. The pink berries are macroscopic, photosynthetic microbial aggregates composed primarily of two closely associated species: sulfide-oxidizing purple sulfur bacteria (PB-PSB1) and sulfate-reducing bacteria (PB-SRB1). Using metagenomic sequencing and (34) S-enriched sulfate stable isotope probing coupled with nanoSIMS, we demonstrate interspecies transfer of reduced sulfur metabolites from PB-SRB1 to PB-PSB1. The pink berries catalyse net sulfide oxidation and maintain internal sulfide concentrations of 0-500 μm. Sulfide within the berries, captured on silver wires and analysed using secondary ion mass spectrometer, increased in abundance towards the berry interior, while δ(34) S-sulfide decreased from 6‰ to -31‰ from the exterior to interior of the berry. These values correspond to sulfate-sulfide isotopic fractionations (15-53‰) consistent with either sulfate reduction or a mixture of reductive and oxidative metabolisms. Together this combined metagenomic and high-resolution isotopic analysis demonstrates active sulfur cycling at the microscale within well-structured macroscopic consortia consisting of sulfide-oxidizing anoxygenic phototrophs and sulfate-reducing bacteria.
© 2014 The Authors. Environmental Microbiology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures


References
-
- Baumgartner LK, Reid RP, Dupraz C, Decho AW, Buckley DH, Spear JR, et al. Sulfate reducing bacteria in microbial mats: changing paradigms, new discoveries. Sediment Geol. 2006;185:131–145.
-
- Baumgartner LK, Dupraz C, Buckley DH, Spear JR, Pace NR. Visscher PT. Microbial species richness and metabolic activities in hypersaline microbial mats: insight into biosignature formation through lithification. Astrobiology. 2009;9:861–874. - PubMed
-
- Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

