T-KDE: a method for genome-wide identification of constitutive protein binding sites from multiple ChIP-seq data sets
- PMID: 24428924
- PMCID: PMC3903014
- DOI: 10.1186/1471-2164-15-27
T-KDE: a method for genome-wide identification of constitutive protein binding sites from multiple ChIP-seq data sets
Abstract
Background: A protein may bind to its target DNA sites constitutively, i.e., regardless of cell type. Intuitively, constitutive binding sites should be biologically functional. A prerequisite for understanding their functional relevance is knowing all their locations for a protein of interest. Genome-wide discovery of constitutive binding sites requires robust and efficient computational methods to integrate results from numerous binding experiments. Such methods are lacking, however.
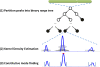
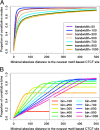
Results: To locate constitutive binding sites for a protein using ChIP-seq data for that protein from multiple cell lines, we developed a method, T-KDE, which combines a binary range tree with a kernel density estimator. Using 132 CTCF (CCCTC-binding factor) ChIP-seq datasets, we showed that the number of constitutive sites identified by T-KDE is robust to the choice of tuning parameter and that T-KDE identifies binding site locations more accurately than a binning approach. Furthermore, T-KDE can identify constitutive sites that are missed by a motif-based approach either because a bound site failed to reach the motif significance cutoff or because the peak sequence scanned was too short. By studying sites declared constitutive by T-KDE but not by the motif-based approach, we discovered two new CTCF motif variants. Using ENCODE data on 22 transcription factors (TF) in 132 cell lines, we identified constitutive binding sites for each TF and provide evidence that, for some TFs, they may be biologically meaningful.
Conclusions: T-KDE is an efficient and effective method to predict constitutive protein binding sites using ChIP-seq peaks from multiple cell lines. Besides constitutive binding sites for a given protein, T-KDE can identify genomic "hot spots" where several different proteins bind and, conversely, cell-type-specific sites bound by a given protein.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

