Sequence-based optimization of a quantitative real-time PCR assay for detection of Plasmodium ovale and Plasmodium malariae
- PMID: 24430459
- PMCID: PMC3993485
- DOI: 10.1128/JCM.03477-13
Sequence-based optimization of a quantitative real-time PCR assay for detection of Plasmodium ovale and Plasmodium malariae
Abstract
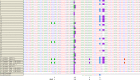
Although microscopic examination of Giemsa-stained blood smears remains the gold standard for the diagnosis of malaria, molecular detection using PCR is becoming increasingly popular. Due to discrepant PCR and microscopy results, we aimed to optimize our detection assays for Plasmodium malariae and Plasmodium ovale by sequencing the 18S rRNA region and developing a new primer and probe set for real-time quantitative PCR (qPCR). Clinical specimens positive for P. malariae (n = 15) or P. ovale (n = 33) underwent amplification and sequencing of the 18S rRNA region. Based on sequence discrepancies between our current primer/probe and clinical isolates, degenerate P. ovale primer and probe were developed to determine if their performance characteristics improved. The reference (gold) standard was microscopy. No 18S sequence heterogeneity was observed among the P. malariae isolates, and the sensitivity and specificity of our current P. malariae qPCR assay were both 100%. Compared to microscopy, the sensitivity and specificity of our current P. ovale qPCR assay were 72.7% and 100%, respectively. Five single nucleotide polymorphisms (SNPs) were identified in P. ovale. The sensitivity of the new P. ovale assay increased to 100% with 100% specificity. We therefore improved the performance characteristics of our P. ovale molecular detection assay through the development of a degenerate primer and probe set which accommodates 18S SNPs among the 2 subspecies of P. ovale. Given the suboptimal sensitivity of rapid diagnostic tests for non-falciparum malaria and the typically low parasitemia of P. malariae and P. ovale, a well-performing confirmatory molecular assay is imperative for clinical laboratories.
Figures
References
-
- Obare P, Ogutu B, Adams M, Odera JS, Lilley K, Dosoo D, Adhiambo C, Owusu-Agyei S, Binka F, Wanja E, Johnson J. 2013. Misclassification of Plasmodium infections by conventional microscopy and the impact of remedial training on the proficiency of laboratory technicians in species identification. Malar. J. 12:113. 10.1186/1475-2875-12-113 - DOI - PMC - PubMed
-
- Khairnar K, Martin D, Lau R, Ralevski F, Pillai DR. 2009. Multiplex real-time quantitative PCR, microscopy and rapid diagnostic immuno-chromatographic tests for the detection of Plasmodium spp: performance, limit of detection analysis and quality assurance. Malar. J. 8:284. 10.1186/1475-2875-8-284 - DOI - PMC - PubMed
-
- Cohen R, Feghali K, Alemayehu S, Komisar J, Hang J, Weina PJ, Coggeshall P, Kamau E, Zapor M. 2013. Use of qPCR and genomic sequencing to diagnose Plasmodium ovale wallikeri malaria in a returned soldier in the setting of a negative rapid diagnostic assay. Am. J. Trop. Med. Hyg. 89:501–506. 10.4269/ajtmh.12-0724 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

