Microbial stratification in low pH oxic and suboxic macroscopic growths along an acid mine drainage
- PMID: 24430486
- PMCID: PMC4030236
- DOI: 10.1038/ismej.2013.242
Microbial stratification in low pH oxic and suboxic macroscopic growths along an acid mine drainage
Abstract
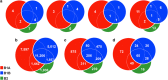
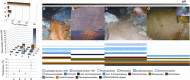
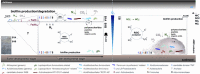
Macroscopic growths at geographically separated acid mine drainages (AMDs) exhibit distinct populations. Yet, local heterogeneities are poorly understood. To gain novel mechanistic insights into this, we used OMICs tools to profile microbial populations coexisting in a single pyrite gallery AMD (pH ∼2) in three distinct compartments: two from a stratified streamer (uppermost oxic and lowermost anoxic sediment-attached strata) and one from a submerged anoxic non-stratified mat biofilm. The communities colonising pyrite and those in the mature formations appear to be populated by the greatest diversity of bacteria and archaea (including 'ARMAN' (archaeal Richmond Mine acidophilic nano-organisms)-related), as compared with the known AMD, with ∼44.9% unclassified sequences. We propose that the thick polymeric matrix may provide a safety shield against the prevailing extreme condition and also a massive carbon source, enabling non-typical acidophiles to develop more easily. Only 1 of 39 species were shared, suggesting a high metabolic heterogeneity in local microenvironments, defined by the O2 concentration, spatial location and biofilm architecture. The suboxic mats, compositionally most similar to each other, are more diverse and active for S, CO2, CH4, fatty acid and lipopolysaccharide metabolism. The oxic stratum of the streamer, displaying a higher diversity of the so-called 'ARMAN'-related Euryarchaeota, shows a higher expression level of proteins involved in signal transduction, cell growth and N, H2, Fe, aromatic amino acids, sphingolipid and peptidoglycan metabolism. Our study is the first to highlight profound taxonomic and functional shifts in single AMD formations, as well as new microbial species and the importance of H2 in acidic suboxic macroscopic growths.
Figures





References
-
- Amils R, González-Toril E, Aguilera A, Rodríguez N, Fernández-Remolar D, Gómez F, et al. From río tinto to Mars: the terrestrial and extraterrestrial ecology of acidophiles. Adv Appl Microbiol. 2011;77:41–70. - PubMed
-
- Baker BJ, Banfield JF. Microbial communities in acid mine drainage. FEMS Microbiol Ecol. 2003;44:139–152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

