An H3K9/S10 methyl-phospho switch modulates Polycomb and Pol II binding at repressed genes during differentiation
- PMID: 24430871
- PMCID: PMC3952858
- DOI: 10.1091/mbc.E13-10-0628
An H3K9/S10 methyl-phospho switch modulates Polycomb and Pol II binding at repressed genes during differentiation
Abstract
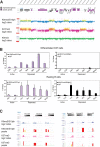
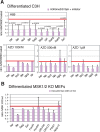
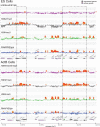
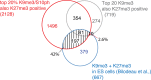
Methylated histones H3K9 and H3K27 are canonical epigenetic silencing modifications in metazoan organisms, but the relationship between the two modifications has not been well characterized. H3K9me3 coexists with H3K27me3 in pluripotent and differentiated cells. However, we find that the functioning of H3K9me3 is altered by H3S10 phosphorylation in differentiated postmitotic osteoblasts and cycling B cells. Deposition of H3K9me3/S10ph at silent genes is partially mediated by the mitogen- and stress-activated kinases (MSK1/2) and the Aurora B kinase. Acquisition of H3K9me3/S10ph during differentiation correlates with loss of paused S5 phosphorylated RNA polymerase II, which is present on Polycomb-regulated genes in embryonic stem cells. Reduction of the levels of H3K9me3/S10ph by kinase inhibition results in increased binding of RNAPIIS5ph and the H3K27 methyltransferase Ezh1 at silent promoters. Our results provide evidence of a novel developmentally regulated methyl-phospho switch that modulates Polycomb regulation in differentiated cells and stabilizes repressed states.
Figures
References
-
- Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. - PubMed
-
- Banfalvi G. Cell cycle synchronization of animal cells and nuclei by centrifugal elutriation. Nat Protoc. 2008;3:663–673. - PubMed
-
- Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006a;125:315–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

