Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates
- PMID: 24431113
- PMCID: PMC4063286
- DOI: 10.1126/scitranslmed.3007361
Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates
Abstract
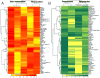
Immunohistochemistry-based clinical diagnoses require invasive core biopsies and use a limited number of protein stains to identify and classify cancers. We introduce a technology that allows analysis of hundreds of proteins from minimally invasive fine-needle aspirates (FNAs), which contain much smaller numbers of cells than core biopsies. The method capitalizes on DNA-barcoded antibody sensing, where barcodes can be photocleaved and digitally detected without any amplification steps. After extensive benchmarking in cell lines, this method showed high reproducibility and achieved single-cell sensitivity. We used this approach to profile ~90 proteins in cells from FNAs and subsequently map patient heterogeneity at the protein level. Additionally, we demonstrate how the method could be used as a clinical tool to identify pathway responses to molecularly targeted drugs and to predict drug response in patient samples. This technique combines specificity with ease of use to offer a new tool for understanding human cancers and designing future clinical trials.
Conflict of interest statement
Figures


References
-
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. published online EpubJun 4 . - DOI - PubMed
-
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. published online EpubAug 2 . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

