MEG and EEG data analysis with MNE-Python
- PMID: 24431986
- PMCID: PMC3872725
- DOI: 10.3389/fnins.2013.00267
MEG and EEG data analysis with MNE-Python
Abstract
Magnetoencephalography and electroencephalography (M/EEG) measure the weak electromagnetic signals generated by neuronal activity in the brain. Using these signals to characterize and locate neural activation in the brain is a challenge that requires expertise in physics, signal processing, statistics, and numerical methods. As part of the MNE software suite, MNE-Python is an open-source software package that addresses this challenge by providing state-of-the-art algorithms implemented in Python that cover multiple methods of data preprocessing, source localization, statistical analysis, and estimation of functional connectivity between distributed brain regions. All algorithms and utility functions are implemented in a consistent manner with well-documented interfaces, enabling users to create M/EEG data analysis pipelines by writing Python scripts. Moreover, MNE-Python is tightly integrated with the core Python libraries for scientific comptutation (NumPy, SciPy) and visualization (matplotlib and Mayavi), as well as the greater neuroimaging ecosystem in Python via the Nibabel package. The code is provided under the new BSD license allowing code reuse, even in commercial products. Although MNE-Python has only been under heavy development for a couple of years, it has rapidly evolved with expanded analysis capabilities and pedagogical tutorials because multiple labs have collaborated during code development to help share best practices. MNE-Python also gives easy access to preprocessed datasets, helping users to get started quickly and facilitating reproducibility of methods by other researchers. Full documentation, including dozens of examples, is available at http://martinos.org/mne.
Keywords: electroencephalography (EEG); magnetoencephalography (MEG); neuroimaging; open-source; python; software.
Figures

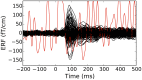
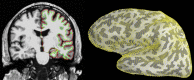
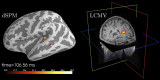
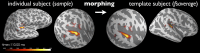




References
-
- Becker R. A., Cleveland W. S., Shyu M.-J., Kaluzny S. P. (1996). A Tour of Trellis Graphics. Technical Report, Bell Laboratories
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 10.1016/j.neuroimage.2012.07.004 - DOI
-
- Buitinck L., Louppe G., Blondel M., Pedregosa F., Mueller A., Grisel O., et al. (2013). API design for machine learning software: experiences from the scikit-learn project. in European Conference on Machine Learning and Principles and Practices of Knowledge Discovery in Databases, Prague
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

