Succession of the turkey gastrointestinal bacterial microbiome related to weight gain
- PMID: 24432198
- PMCID: PMC3883494
- DOI: 10.7717/peerj.237
Succession of the turkey gastrointestinal bacterial microbiome related to weight gain
Abstract
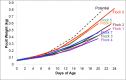
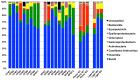
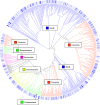
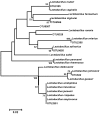
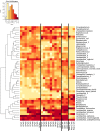
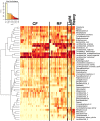
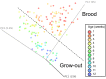
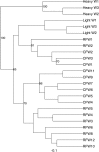
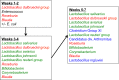
Because of concerns related to the use of antibiotics in animal agriculture, antibiotic-free alternatives are greatly needed to prevent disease and promote animal growth. One of the current challenges facing commercial turkey production in Minnesota is difficulty obtaining flock average weights typical of the industry standard, and this condition has been coined "Light Turkey Syndrome" or LTS. This condition has been identified in Minnesota turkey flocks for at least five years, and it has been observed that average flock body weights never approach their genetic potential. However, a single causative agent responsible for these weight reductions has not been identified despite numerous efforts to do so. The purpose of this study was to identify the bacterial community composition within the small intestines of heavy and light turkey flocks using 16S rRNA sequencing, and to identify possible correlations between microbiome and average flock weight. This study also sought to define the temporal succession of bacteria occurring in the turkey ileum. Based upon 2.7 million sequences across nine different turkey flocks, dominant operational taxonomic units (OTUs) were identified and compared between the flocks studied. OTUs that were associated with heavier weight flocks included those with similarity to Candidatus division Arthromitus and Clostridium bartlettii, while these flocks had decreased counts of several Lactobacillus species compared to lighter weight flocks. The core bacterial microbiome succession in commercial turkeys was also defined. Several defining markers of microbiome succession were identified, including the presence or abundance of Candidatus division Arthromitus, Lactobacillus aviarius, Lactobacillus ingluviei, Lactobacillus salivarius, and Clostridium bartlettii. Overall, the succession of the ileum bacterial microbiome in commercial turkeys proceeds in a predictable manner. Efforts to prevent disease and promote growth in the absence of antibiotics could involve target dominant bacteria identified in the turkey ileum that are associated with increased weight gain.
Keywords: 16S; Bacteria; Candidatus Arthromitus; Gastrointestinal; Lactobacillus; Microbiome; Poultry; Segmented filamentous bacteria; Succession; Turkeys.
Figures
References
-
- Bartram AK, Lynch MD, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Applied and Environmental Microbiology. 2011;77:3846–3852. doi: 10.1128/AEM.02772-10. - DOI - PMC - PubMed
-
- Calvert A. Master Thesis. Department of Animal Sciences. 2012. Light turkey syndrome: field study and inoculation trial.
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

