Release of protein A from the cell wall of Staphylococcus aureus
- PMID: 24434550
- PMCID: PMC3910568
- DOI: 10.1073/pnas.1317181111
Release of protein A from the cell wall of Staphylococcus aureus
Abstract
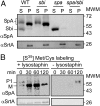
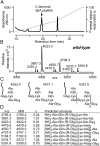
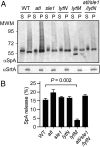
Staphylococcal protein A (SpA) is anchored to the cell wall envelope of Staphylococcus aureus by sortase A, which links the threonyl (T) of its C-terminal LPXTG motif to peptidoglycan cross-bridges (i.e., Gly5). SpA binds the Fcγ domains of IgG and protects staphylococci from opsonophagocytic clearance. Moreover, SpA cross-links B-cell receptors to modify host adaptive immune responses. The mechanisms whereby SpA is released from the bacterial surface to access the host's immune system are not known. Here we demonstrate that SpA is released with murein tetrapeptide-tetraglycyl [L-Ala-D-iGln-(SpA-Gly5)L-Lys-D-Ala-Gly4] linked to its C-terminal threonyl. LytN, a cross-wall murein hydrolase, contributes to the release of SpA by removing amino sugars [i.e., N-acetylmuramic acid-N-acetylglucosamine (MurNAc-GlcNAc)] from attached peptidoglycan, whereas LytM, a pentaglycyl-endopeptidase, triggers polypeptide release from the bacterial envelope. A model is proposed whereby murein hydrolases cleave the anchor structure of released SpA to modify host immune responses.
Keywords: Gram-positive bacteria; sortase-anchored protein; surface protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Anchor structure of staphylococcal surface proteins. II. Cooh-terminal structure of muramidase and amidase-solubilized surface protein.J Biol Chem. 1998 Oct 30;273(44):29135-42. doi: 10.1074/jbc.273.44.29135. J Biol Chem. 1998. PMID: 9786922
-
Anchor structure of staphylococcal surface proteins. III. Role of the FemA, FemB, and FemX factors in anchoring surface proteins to the bacterial cell wall.J Biol Chem. 1998 Oct 30;273(44):29143-9. doi: 10.1074/jbc.273.44.29143. J Biol Chem. 1998. PMID: 9786923
-
Anchor structure of staphylococcal surface proteins. A branched peptide that links the carboxyl terminus of proteins to the cell wall.J Biol Chem. 1997 Aug 29;272(35):22285-92. doi: 10.1074/jbc.272.35.22285. J Biol Chem. 1997. PMID: 9268378
-
Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus.Mol Microbiol. 2001 Jun;40(5):1049-57. doi: 10.1046/j.1365-2958.2001.02411.x. Mol Microbiol. 2001. PMID: 11401711 Review.
-
Staphylococcal Protein Secretion and Envelope Assembly.Microbiol Spectr. 2019 Jul;7(4):10.1128/microbiolspec.gpp3-0070-2019. doi: 10.1128/microbiolspec.GPP3-0070-2019. Microbiol Spectr. 2019. PMID: 31267890 Free PMC article. Review.
Cited by
-
The Staphylococcal Biofilm: Adhesins, Regulation, and Host Response.Microbiol Spectr. 2016 Apr;4(2):10.1128/microbiolspec.VMBF-0022-2015. doi: 10.1128/microbiolspec.VMBF-0022-2015. Microbiol Spectr. 2016. PMID: 27227309 Free PMC article. Review.
-
Characterization of recombinant wild-type and nontoxigenic protein A from Staphylococcus pseudintermedius.Virulence. 2018;9(1):1050-1061. doi: 10.1080/21505594.2018.1489199. Virulence. 2018. PMID: 30052123 Free PMC article.
-
Using Knock-Out Mutants to Investigate the Adhesion of Staphylococcus aureus to Abiotic Surfaces.Int J Mol Sci. 2021 Nov 4;22(21):11952. doi: 10.3390/ijms222111952. Int J Mol Sci. 2021. PMID: 34769382 Free PMC article.
-
Looking through Staphylococcus pseudintermedius infections: Could SpA be considered a possible vaccine target?Virulence. 2018 Dec 31;9(1):703-706. doi: 10.1080/21505594.2018.1426964. Virulence. 2018. PMID: 29457988 Free PMC article. No abstract available.
-
The effect of Staphylococcus aureus on innate and adaptive immunity and potential immunotherapy for S. aureus-induced osteomyelitis.Front Immunol. 2023 Sep 8;14:1219895. doi: 10.3389/fimmu.2023.1219895. eCollection 2023. Front Immunol. 2023. PMID: 37744377 Free PMC article. Review.
References
-
- Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. - PubMed
-
- Strominger JL, Izaki K, Matsuhashi M, Tipper DJ. Peptidoglycan transpeptidase and D-alanine carboxypeptidase: Penicillin-sensitive enzymatic reactions. Fed Proc. 1967;26(1):9–22. - PubMed
-
- Park JT, Strominger JL. Mode of action of penicillin. Science. 1957;125(3238):99–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

