Synthetic biology in mammalian cells: next generation research tools and therapeutics
- PMID: 24434884
- PMCID: PMC4032074
- DOI: 10.1038/nrm3738
Synthetic biology in mammalian cells: next generation research tools and therapeutics
Abstract
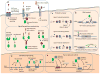
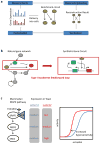
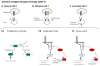
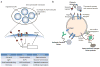
Recent progress in DNA manipulation and gene circuit engineering has greatly improved our ability to programme and probe mammalian cell behaviour. These advances have led to a new generation of synthetic biology research tools and potential therapeutic applications. Programmable DNA-binding domains and RNA regulators are leading to unprecedented control of gene expression and elucidation of gene function. Rebuilding complex biological circuits such as T cell receptor signalling in isolation from their natural context has deepened our understanding of network motifs and signalling pathways. Synthetic biology is also leading to innovative therapeutic interventions based on cell-based therapies, protein drugs, vaccines and gene therapies.
Figures


References
-
- Endy D. Foundations for engineering biology. Nature. 2005;438:449–53. - PubMed
-
- Knight T. DARPA BioComp Plasmid Distribution 1.00 of Standard Biobrick Components. MIT Synthetic Biology Working Group Reports. 2002
-
- Mutalik VK, et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods. 2013;10:354–60. - PubMed
-
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–42. - PubMed
-
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

