Gα12 structural determinants of Hsp90 interaction are necessary for serum response element-mediated transcriptional activation
- PMID: 24435554
- PMCID: PMC3965892
- DOI: 10.1124/mol.113.088443
Gα12 structural determinants of Hsp90 interaction are necessary for serum response element-mediated transcriptional activation
Abstract
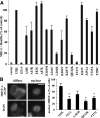
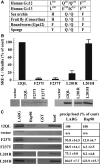
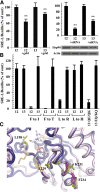
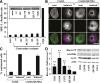
The G12/13 class of heterotrimeric G proteins, comprising the α-subunits Gα12 and Gα13, regulates multiple aspects of cellular behavior, including proliferation and cytoskeletal rearrangements. Although guanine nucleotide exchange factors for the monomeric G protein Rho (RhoGEFs) are well characterized as effectors of this G protein class, a variety of other downstream targets has been reported. To identify Gα12 determinants that mediate specific protein interactions, we used a structural and evolutionary comparison between the G12/13, Gs, Gi, and Gq classes to identify "class-distinctive" residues in Gα12 and Gα13. Mutation of these residues in Gα12 to their deduced ancestral forms revealed a subset necessary for activation of serum response element (SRE)-mediated transcription, a G12/13-stimulated pathway implicated in cell proliferative signaling. Unexpectedly, this subset of Gα12 mutants showed impaired binding to heat-shock protein 90 (Hsp90) while retaining binding to RhoGEFs. Corresponding mutants of Gα13 exhibited robust SRE activation, suggesting a Gα12-specific mechanism, and inhibition of Hsp90 by geldanamycin or small interfering RNA-mediated lowering of Hsp90 levels resulted in greater downregulation of Gα12 than Gα13 signaling in SRE activation experiments. Furthermore, the Drosophila G12/13 homolog Concertina was unable to signal to SRE in mammalian cells, and Gα12:Concertina chimeras revealed Gα12-specific determinants of SRE activation within the switch regions and a C-terminal region. These findings identify Gα12 determinants of SRE activation, implicate Gα12:Hsp90 interaction in this signaling mechanism, and illuminate structural features that arose during evolution of Gα12 and Gα13 to allow bifurcated mechanisms of signaling to a common cell proliferative pathway.
Figures


References
-
- Adarichev VA, Vaiskunaite R, Niu J, Balyasnikova IV, Voyno-Yasenetskaya TA. (2003) Gα13-mediated transformation and apoptosis are permissively dependent on basal ERK activity. Am J Physiol Cell Physiol 285:C922–C934 - PubMed
-
- Barrett K, Leptin M, Settleman J. (1997) The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell 91:905–915 - PubMed
-
- Bhattacharyya R, Wedegaertner PB. (2000) Galpha 13 requires palmitoylation for plasma membrane localization, Rho-dependent signaling, and promotion of p115-RhoGEF membrane binding. J Biol Chem 275:14992–14999 - PubMed
-
- Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. (2003) Insights into G protein structure, function, and regulation. Endocr Rev 24:765–781 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

