Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution
- PMID: 24436049
- PMCID: PMC4293454
- DOI: 10.1158/2159-8290.CD-13-0285
Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution
Abstract
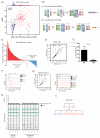
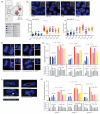
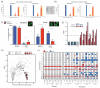
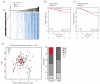
The contribution of whole-genome doubling to chromosomal instability (CIN) and tumor evolution is unclear. We use long-term culture of isogenic tetraploid cells from a stable diploid colon cancer progenitor to investigate how a genome-doubling event affects genome stability over time. Rare cells that survive genome doubling demonstrate increased tolerance to chromosome aberrations. Tetraploid cells do not exhibit increased frequencies of structural or numerical CIN per chromosome. However, the tolerant phenotype in tetraploid cells, coupled with a doubling of chromosome aberrations per cell, allows chromosome abnormalities to evolve specifically in tetraploids, recapitulating chromosomal changes in genomically complex colorectal tumors. Finally, a genome-doubling event is independently predictive of poor relapse-free survival in early-stage disease in two independent cohorts in multivariate analyses [discovery data: hazard ratio (HR), 4.70, 95% confidence interval (CI), 1.04-21.37; validation data: HR, 1.59, 95% CI, 1.05-2.42]. These data highlight an important role for the tolerance of genome doubling in driving cancer genome evolution.
Significance: Our work sheds light on the importance of whole-genome–doubling events in colorectal cancer evolution. We show that tetraploid cells undergo rapid genomic changes and recapitulate the genetic alterations seen in chromosomally unstable tumors. Furthermore, we demonstrate that a genome-doubling event is prognostic of poor relapse-free survival in this disease type.
2014 AACR
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

