Suppression of microRNA-9 by mutant EGFR signaling upregulates FOXP1 to enhance glioblastoma tumorigenicity
- PMID: 24436148
- PMCID: PMC3947420
- DOI: 10.1158/0008-5472.CAN-13-2117
Suppression of microRNA-9 by mutant EGFR signaling upregulates FOXP1 to enhance glioblastoma tumorigenicity
Abstract
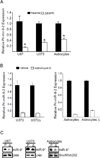
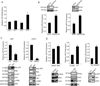
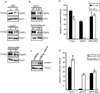
The EGF receptor (EGFR) is amplified and mutated in glioblastoma, in which its common mutation (ΔEGFR, also called EGFRvIII) has a variety of activities that promote growth and inhibit death, thereby conferring a strong tumor-enhancing effect. This range of activities suggested to us that ΔEGFR might exert its influence through pleiotropic effectors, and we hypothesized that microRNAs might serve such a function. Here, we report that ΔEGFR specifically suppresses one such microRNA, namely miR-9, through the Ras/PI3K/AKT axis that it is known to activate. Correspondingly, expression of miR-9 antagonizes the tumor growth advantage conferred by ΔEGFR. Silencing of FOXP1, a miR-9 target, inhibits ΔEGFR-dependent tumor growth and, conversely, de-repression of FOXP1, as a consequence of miR-9 inhibition, increases tumorigenicity. FOXP1 was sufficient to increase tumor growth in the absence of oncogenic ΔEGFR signaling. The significance of these findings is underscored by our finding that high FOXP1 expression predicts poor survival in a cohort of 131 patients with glioblastoma. Collectively, these data suggest a novel regulatory mechanism by which ΔEGFR suppression of miR-9 upregulates FOXP1 to increase tumorigenicity.
©2014 AACR
Conflict of interest statement
No potential conflicts of interest were disclosed by the authors.
Figures


Similar articles
-
Regulation of HGF expression by ΔEGFR-mediated c-Met activation in glioblastoma cells.Neoplasia. 2013 Jan;15(1):73-84. doi: 10.1593/neo.121536. Neoplasia. 2013. PMID: 23359207 Free PMC article.
-
Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases.Proc Natl Acad Sci U S A. 1998 May 12;95(10):5724-9. doi: 10.1073/pnas.95.10.5724. Proc Natl Acad Sci U S A. 1998. PMID: 9576951 Free PMC article.
-
Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas.Cancer Res. 2002 Nov 15;62(22):6764-9. Cancer Res. 2002. PMID: 12438278
-
Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop.J Exp Clin Cancer Res. 2020 Sep 22;39(1):196. doi: 10.1186/s13046-020-01695-8. J Exp Clin Cancer Res. 2020. PMID: 32962742 Free PMC article.
-
Nuclear EGFRvIII-STAT5b complex contributes to glioblastoma cell survival by direct activation of the Bcl-XL promoter.Int J Cancer. 2013 Feb 1;132(3):509-20. doi: 10.1002/ijc.27690. Epub 2012 Jul 9. Int J Cancer. 2013. PMID: 22729867 Free PMC article.
Cited by
-
FOXP1 potentiates Wnt/β-catenin signaling in diffuse large B cell lymphoma.Sci Signal. 2015 Feb 3;8(362):ra12. doi: 10.1126/scisignal.2005654. Sci Signal. 2015. PMID: 25650440 Free PMC article.
-
Genetic Abnormalities, Clonal Evolution, and Cancer Stem Cells of Brain Tumors.Med Sci (Basel). 2018 Oct 2;6(4):85. doi: 10.3390/medsci6040085. Med Sci (Basel). 2018. PMID: 30279357 Free PMC article. Review.
-
MiR-206 is down-regulated and suppresses cell proliferation by targeting FOXP1 in brain gliomas.Int J Clin Exp Pathol. 2018 Jul 1;11(7):3405-3415. eCollection 2018. Int J Clin Exp Pathol. 2018. PMID: 31949718 Free PMC article.
-
PUMILIO/FOXP1 signaling drives expansion of hematopoietic stem/progenitor and leukemia cells.Blood. 2017 May 4;129(18):2493-2506. doi: 10.1182/blood-2016-10-747436. Epub 2017 Feb 23. Blood. 2017. PMID: 28232582 Free PMC article.
-
MiR-9 Promotes Apoptosis Via Suppressing SMC1A Expression in GBM Cell Lines.Curr Chem Genom Transl Med. 2017 Jul 31;11:31-40. doi: 10.2174/2213988501711010031. eCollection 2017. Curr Chem Genom Transl Med. 2017. PMID: 28868238 Free PMC article.
References
-
- Nishikawa R, Sugiyama T, Narita Y, Furnari F, Cavenee WK, Matsutani M. Immunohistochemical analysis of the mutant epidermal growth factor, deltaEGFR, in glioblastoma. Brain Tumor Pathol. 2004;21:53–56. - PubMed
-
- Lammering G, Valerie K, Lin PS, Hewit TH, Schmidt-Ullrich RK. Radiation-induced activation of a common variant of EGFR confers enhanced radioresistance. Radiother Oncol. 2004;72:267–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

