DNA adductomics
- PMID: 24437709
- PMCID: PMC3997222
- DOI: 10.1021/tx4004352
DNA adductomics
Abstract
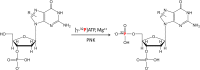
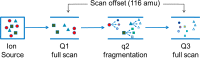
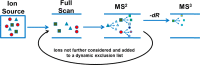
Systems toxicology is a broad-based approach to describe many of the toxicological features that occur within a living system under stress or subjected to exogenous or endogenous exposures. The ultimate goal is to capture an overview of all exposures and the ensuing biological responses of the body. The term exposome has been employed to refer to the totality of all exposures, and systems toxicology investigates how the exposome influences health effects and consequences of exposures over a lifetime. The tools to advance systems toxicology include high-throughput transcriptomics, proteomics, metabolomics, and adductomics, which is still in its infancy. A well-established methodology for the comprehensive measurement of DNA damage resulting from every day exposures is not fully developed. During the past several decades, the (32)P-postlabeling technique has been employed to screen the damage to DNA induced by multiple classes of genotoxicants; however, more robust, specific, and quantitative methods have been sought to identify and quantify DNA adducts. Although triple quadrupole and ion trap mass spectrometry, particularly when using multistage scanning (LC-MS(n)), have shown promise in the field of DNA adductomics, it is anticipated that high-resolution and accurate-mass LC-MS(n) instrumentation will play a major role in assessing global DNA damage. Targeted adductomics should also benefit greatly from improved triple quadrupole technology. Once the analytical MS methods are fully mature, DNA adductomics along with other -omics tools will contribute greatly to the field of systems toxicology.
Figures




References
-
- Waters M. D.; Fostel J. M. (2004) Toxicogenomics and systems toxicology: Aims and prospects. Nat. Rev. Genet. 5, 936–948. - PubMed
-
- Heijne W. H.; Kienhuis A. S.; van Ommen B.; Stierum R. H.; Groten J. P. (2005) Systems toxicology: Applications of toxicogenomics, transcriptomics, proteomics and metabolomics in toxicology. Expert Rev. Proteomics 2, 767–780. - PubMed
-
- Craig A.; Sidaway J.; Holmes E.; Orton T.; Jackson D.; Rowlinson R.; Nickson J.; Tonge R.; Wilson I.; Nicholson J. (2006) Systems toxicology: integrated genomic, proteomic and metabonomic analysis of methapyrilene induced hepatotoxicity in the rat. J. Proteome Res. 5, 1586–1601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

