Reversing DNA methylation: mechanisms, genomics, and biological functions
- PMID: 24439369
- PMCID: PMC3938284
- DOI: 10.1016/j.cell.2013.12.019
Reversing DNA methylation: mechanisms, genomics, and biological functions
Abstract
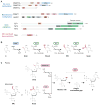
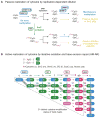
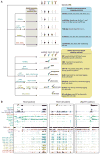
Methylation of cytosines in the mammalian genome represents a key epigenetic modification and is dynamically regulated during development. Compelling evidence now suggests that dynamic regulation of DNA methylation is mainly achieved through a cyclic enzymatic cascade comprised of cytosine methylation, iterative oxidation of methyl group by TET dioxygenases, and restoration of unmodified cytosines by either replication-dependent dilution or DNA glycosylase-initiated base excision repair. In this review, we discuss the mechanism and function of DNA demethylation in mammalian genomes, focusing particularly on how developmental modulation of the cytosine-modifying pathway is coupled to active reversal of DNA methylation in diverse biological processes.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


References
-
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

