Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events
- PMID: 24439375
- PMCID: PMC3909490
- DOI: 10.1016/j.cell.2013.11.047
Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events
Abstract
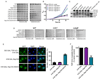
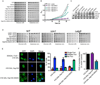
There are no therapies that reverse the proteotoxic misfolding events that underpin fatal neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS) and Parkinson's disease (PD). Hsp104, a conserved hexameric AAA+ protein from yeast, solubilizes disordered aggregates and amyloid but has no metazoan homolog and only limited activity against human neurodegenerative disease proteins. Here, we reprogram Hsp104 to rescue TDP-43, FUS, and α-synuclein proteotoxicity by mutating single residues in helix 1, 2, or 3 of the middle domain or the small domain of nucleotide-binding domain 1. Potentiated Hsp104 variants enhance aggregate dissolution, restore proper protein localization, suppress proteotoxicity, and in a C. elegans PD model attenuate dopaminergic neurodegeneration. Potentiating mutations reconfigure how Hsp104 subunits collaborate, desensitize Hsp104 to inhibition, obviate any requirement for Hsp70, and enhance ATPase, translocation, and unfoldase activity. Our work establishes that disease-associated aggregates and amyloid are tractable targets and that enhanced disaggregases can restore proteostasis and mitigate neurodegeneration.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins.Dis Model Mech. 2014 Oct;7(10):1175-84. doi: 10.1242/dmm.016113. Epub 2014 Jul 25. Dis Model Mech. 2014. PMID: 25062688 Free PMC article.
-
Therapeutic genetic variation revealed in diverse Hsp104 homologs.Elife. 2020 Dec 15;9:e57457. doi: 10.7554/eLife.57457. Elife. 2020. PMID: 33319748 Free PMC article.
-
Isolating potentiated Hsp104 variants using yeast proteinopathy models.J Vis Exp. 2014 Nov 11;(93):e52089. doi: 10.3791/52089. J Vis Exp. 2014. PMID: 25407485 Free PMC article.
-
Engineering therapeutic protein disaggregases.Mol Biol Cell. 2016 May 15;27(10):1556-60. doi: 10.1091/mbc.E15-10-0693. Mol Biol Cell. 2016. PMID: 27255695 Free PMC article. Review.
-
Engineering enhanced protein disaggregases for neurodegenerative disease.Prion. 2015;9(2):90-109. doi: 10.1080/19336896.2015.1020277. Prion. 2015. PMID: 25738979 Free PMC article. Review.
Cited by
-
Solid-to-liquid phase transition in the dissolution of cytosolic misfolded-protein aggregates.iScience. 2023 Oct 28;26(12):108334. doi: 10.1016/j.isci.2023.108334. eCollection 2023 Dec 15. iScience. 2023. PMID: 38025775 Free PMC article.
-
Modifier pathways in polyglutamine (PolyQ) diseases: from genetic screens to drug targets.Cell Mol Life Sci. 2022 May 3;79(5):274. doi: 10.1007/s00018-022-04280-8. Cell Mol Life Sci. 2022. PMID: 35503478 Free PMC article. Review.
-
Hsp104 N-terminal domain interaction with substrates plays a regulatory role in protein disaggregation.FEBS J. 2022 Sep;289(17):5359-5377. doi: 10.1111/febs.16441. Epub 2022 Mar 30. FEBS J. 2022. PMID: 35305079 Free PMC article.
-
Differential roles for DNAJ isoforms in HTT-polyQ and FUS aggregation modulation revealed by chaperone screens.Nat Commun. 2022 Jan 26;13(1):516. doi: 10.1038/s41467-022-27982-w. Nat Commun. 2022. PMID: 35082301 Free PMC article.
-
Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins.Dis Model Mech. 2014 Oct;7(10):1175-84. doi: 10.1242/dmm.016113. Epub 2014 Jul 25. Dis Model Mech. 2014. PMID: 25062688 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R21HD074510/HD/NICHD NIH HHS/United States
- R01GM099836/GM/NIGMS NIH HHS/United States
- R15 NS075684/NS/NINDS NIH HHS/United States
- R21 HD074510/HD/NICHD NIH HHS/United States
- R01 GM099836/GM/NIGMS NIH HHS/United States
- R21NS067354/NS/NINDS NIH HHS/United States
- R21 NS067354/NS/NINDS NIH HHS/United States
- T32 GM071339/GM/NIGMS NIH HHS/United States
- T32GM071339/GM/NIGMS NIH HHS/United States
- T32 GM007223/GM/NIGMS NIH HHS/United States
- F31 NS079009/NS/NINDS NIH HHS/United States
- F31NS079009/NS/NINDS NIH HHS/United States
- DP2 OD002177/OD/NIH HHS/United States
- R15NS075684/NS/NINDS NIH HHS/United States
- DP2OD002177/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

