Current progress in antiviral strategies
- PMID: 24439476
- PMCID: PMC7112804
- DOI: 10.1016/j.tips.2013.11.006
Current progress in antiviral strategies
Abstract
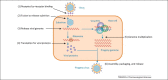
The prevalence of chronic viral infectious diseases, such as human immunodeficiency virus (HIV), hepatitis C virus (HCV), and influenza virus; the emergence and re-emergence of new viral infections, such as picornaviruses and coronaviruses; and, particularly, resistance to currently used antiviral drugs have led to increased demand for new antiviral strategies and reagents. Increased understanding of the molecular mechanisms of viral infection has provided great potential for the discovery of new antiviral agents that target viral proteins or host factors. Virus-targeting antivirals can function directly or indirectly to inhibit the biological functions of viral proteins, mostly enzymatic activities, or to block viral replication machinery. Host-targeting antivirals target the host proteins that are involved in the viral life cycle, regulating the function of the immune system or other cellular processes in host cells. Here we review key targets and considerations for the development of both antiviral strategies.
Keywords: direct virus-targeting antiviral; host-targeting antiviral; indirect virus-targeting antiviral.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures




References
-
- Lyczak J.B., Morrison S.L. Biological and pharmacokinetic properties of a novel immunoglobulin–CD4 fusion protein. Arch. Virol. 1994;139:189–196. - PubMed
-
- Schooley R.T. Recombinant soluble CD4 therapy in patients with the acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. A Phase I–II escalating dosage trial. Ann. Intern. Med. 1990;112:247–253. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

