Trafficking of endosomal Toll-like receptors
- PMID: 24439965
- PMCID: PMC4037363
- DOI: 10.1016/j.tcb.2013.12.002
Trafficking of endosomal Toll-like receptors
Abstract
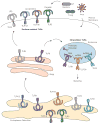
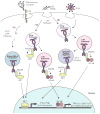
Over the past decade we have learned much about nucleic acid recognition by the innate immune system and in particular by Toll-like receptors (TLRs). These receptors localize to endosomal compartments where they are poised to recognize microbial nucleic acids. Multiple regulatory mechanisms function to limit responses to self DNA or RNA, and breakdowns in these mechanisms can contribute to autoimmune or inflammatory disorders. In this review we discuss our current understanding of the cell biology of TLRs involved in nucleic acid recognition and how localization and trafficking of these receptors regulates their function.
Keywords: AP complex; UNC93B1; autoimmunity; innate immunity; type I interferon.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
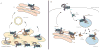
References
-
- Janeway CA. Approaching the Asymptote? Evolution and Revolution in Immunology. Cold Spring Harbor Symposia on Quantitative Biology. 1989;54(0):1–13. - PubMed
-
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature Publishing Group. 2010;11(5):373–384. - PubMed
-
- Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Annals of the New York Academy of Sciences. 2008;1143:1–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

