Characterization of intact N- and O-linked glycopeptides using higher energy collisional dissociation
- PMID: 24440233
- PMCID: PMC5627992
- DOI: 10.1016/j.ab.2014.01.003
Characterization of intact N- and O-linked glycopeptides using higher energy collisional dissociation
Abstract
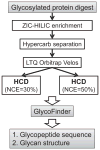
Simultaneous elucidation of the glycan structure and the glycosylation site are needed to reveal the biological function of protein glycosylation. In this study, we employed a recent type of fragmentation termed higher energy collisional dissociation (HCD) to examine fragmentation patterns of intact glycopeptides generated from a mixture of standard glycosylated proteins. The normalized collisional energy (NCE) value for HCD was varied from 30 to 60% to evaluate the optimal conditions for the fragmentation of peptide backbones and glycoconjugates. Our results indicated that HCD with lower NCE values preferentially fragmented the sugar chains attached to the peptides to generate a ladder of neutral loss of monosaccharides, thereby enabling the putative glycan structure characterization. In addition, detection of the oxonium ions enabled unambiguous differentiation of glycopeptides from non-glycopeptides. In contrast, HCD with higher NCE values preferentially fragmented the peptide backbone and, thus, provided information needed for confident peptide identification. We evaluated the HCD approach with alternating NCE parameters for confident characterization of intact N- and O-linked glycopeptides in a single liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. In addition, we applied a novel data analysis pipeline, so-called GlycoFinder, to form a basis for automated data analysis. Overall, 38 unique intact glycopeptides corresponding to eight glycosylation sites (six N-linked and two O-linked sites) were confidently identified from a standard protein mixture. This approach provided concurrent characterization of both the peptide and the glycan, thereby enabling comprehensive structural characterization of glycoproteins in a single LC-MS/MS analysis.
Keywords: Automated identification; Glycopeptides; Glycosylation; HCD; LC–MS/MS; NCE.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni.Mol Cell Proteomics. 2011 Feb;10(2):M000031-MCP201. doi: 10.1074/mcp.M000031-MCP201. Epub 2010 Apr 1. Mol Cell Proteomics. 2011. PMID: 20360033 Free PMC article.
-
Characterization of glycopeptides using a stepped higher-energy C-trap dissociation approach on a hybrid quadrupole orbitrap.Rapid Commun Mass Spectrom. 2018 Aug 30;32(16):1353-1362. doi: 10.1002/rcm.8191. Rapid Commun Mass Spectrom. 2018. PMID: 29873418
-
GPQuest: A Spectral Library Matching Algorithm for Site-Specific Assignment of Tandem Mass Spectra to Intact N-glycopeptides.Anal Chem. 2015;87(10):5181-8. doi: 10.1021/acs.analchem.5b00024. Epub 2015 May 6. Anal Chem. 2015. PMID: 25945896 Free PMC article.
-
Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides.Glycoconj J. 2016 Jun;33(3):261-72. doi: 10.1007/s10719-016-9649-3. Epub 2016 Jan 18. Glycoconj J. 2016. PMID: 26780731 Review.
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
Cited by
-
N-Linked Glycopeptide Identification Based on Open Mass Spectral Library Search.Biomed Res Int. 2018 Aug 14;2018:1564136. doi: 10.1155/2018/1564136. eCollection 2018. Biomed Res Int. 2018. PMID: 30186849 Free PMC article.
-
Maturing Glycoproteomics Technologies Provide Unique Structural Insights into the N-glycoproteome and Its Regulation in Health and Disease.Mol Cell Proteomics. 2016 Jun;15(6):1773-90. doi: 10.1074/mcp.O115.057638. Epub 2016 Feb 29. Mol Cell Proteomics. 2016. PMID: 26929216 Free PMC article. Review.
-
Data-independent oxonium ion profiling of multi-glycosylated biotherapeutics.MAbs. 2018 Oct;10(7):968-978. doi: 10.1080/19420862.2018.1494106. Epub 2018 Aug 1. MAbs. 2018. PMID: 30067433 Free PMC article.
-
Comprehensive Analysis of Protein Glycation Reveals Its Potential Impacts on Protein Degradation and Gene Expression in Human Cells.J Am Soc Mass Spectrom. 2019 Dec;30(12):2480-2490. doi: 10.1007/s13361-019-02197-4. Epub 2019 May 9. J Am Soc Mass Spectrom. 2019. PMID: 31073893 Free PMC article.
-
The Art of Destruction: Optimizing Collision Energies in Quadrupole-Time of Flight (Q-TOF) Instruments for Glycopeptide-Based Glycoproteomics.J Am Soc Mass Spectrom. 2016 Mar;27(3):507-19. doi: 10.1007/s13361-015-1308-6. Epub 2016 Jan 4. J Am Soc Mass Spectrom. 2016. PMID: 26729457 Free PMC article.
References
-
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. - PubMed
-
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. - PubMed
-
- Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. - PubMed
-
- Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–2376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

