Compartmentalization and organelle formation in bacteria
- PMID: 24440431
- PMCID: PMC4318566
- DOI: 10.1016/j.ceb.2013.12.007
Compartmentalization and organelle formation in bacteria
Abstract
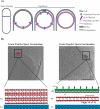
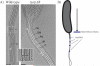
A number of bacterial species rely on compartmentalization to gain specific functionalities that will provide them with a selective advantage. Here, we will highlight several of these modes of bacterial compartmentalization with an eye toward describing the mechanisms of their formation and their evolutionary origins. Spore formation in Bacillus subtilis, outer membrane biogenesis in Gram-negative bacteria and protein diffusion barriers of Caulobacter crescentus will be used to demonstrate the physical, chemical, and compositional remodeling events that lead to compartmentalization. In addition, magnetosomes and carboxysomes will serve as models to examine the interplay between cytoskeletal systems and the subcellular positioning of organelles.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

