The atypical ubiquitin ligase RNF31 stabilizes estrogen receptor α and modulates estrogen-stimulated breast cancer cell proliferation
- PMID: 24441041
- PMCID: PMC4141304
- DOI: 10.1038/onc.2013.573
The atypical ubiquitin ligase RNF31 stabilizes estrogen receptor α and modulates estrogen-stimulated breast cancer cell proliferation
Erratum in
-
Correction: The atypical ubiquitin ligase RNF31 stabilizes estrogen receptor α and modulates estrogen-stimulated breast cancer cell proliferation.Oncogene. 2019 Jan;38(2):299-300. doi: 10.1038/s41388-018-0502-y. Oncogene. 2019. PMID: 30262864 Free PMC article.
Abstract
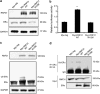
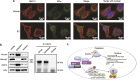
Estrogen receptor α (ERα) is initially expressed in the majority of breast cancers and promotes estrogen-dependent cancer progression by regulating the transcription of genes linked to cell proliferation. ERα status is of clinical importance, as ERα-positive breast cancers can be successfully treated by adjuvant therapy with antiestrogens or aromatase inhibitors. Complications arise from the frequent development of drug resistance that might be caused by multiple alterations, including components of ERα signaling, during tumor progression and metastasis. Therefore, insights into the molecular mechanisms that control ERα expression and stability are of utmost importance to improve breast cancer diagnostics and therapeutics. Here we report that the atypical E3 ubiquitin ligase RNF31 stabilizes ERα and facilitates ERα-stimulated proliferation in breast cancer cell lines. We show that depletion of RNF31 decreases the number of cells in the S phase and reduces the levels of ERα and its downstream target genes, including cyclin D1 and c-myc. Analysis of data from clinical samples confirms correlation between RNF31 expression and the expression of ERα target genes. Immunoprecipitation indicates that RNF31 associates with ERα and increases its stability and mono-ubiquitination, dependent on the ubiquitin ligase activity of RNF31. Our data suggest that association of RNF31 and ERα occurs mainly in the cytosol, consistent with the lack of RNF31 recruitment to ERα-occupied promoters. In conclusion, our study establishes a non-genomic mechanism by which RNF31 via stabilizing ERα levels controls the transcription of estrogen-dependent genes linked to breast cancer cell proliferation.
Figures


Similar articles
-
The ubiquitin ligase RNF181 stabilizes ERα and modulates breast cancer progression.Oncogene. 2020 Oct;39(44):6776-6788. doi: 10.1038/s41388-020-01464-z. Epub 2020 Sep 24. Oncogene. 2020. PMID: 32973333 Free PMC article.
-
Atypical ubiquitin ligase RNF31: the nuclear factor modulator in breast cancer progression.BMC Cancer. 2016 Jul 26;16:538. doi: 10.1186/s12885-016-2575-8. BMC Cancer. 2016. PMID: 27460922 Free PMC article. Review.
-
The GPER1/SPOP axis mediates ubiquitination-dependent degradation of ERα to inhibit the growth of breast cancer induced by oestrogen.Cancer Lett. 2021 Feb 1;498:54-69. doi: 10.1016/j.canlet.2020.10.019. Epub 2020 Oct 15. Cancer Lett. 2021. PMID: 33069770
-
SMURF1 facilitates estrogen receptor ɑ signaling in breast cancer cells.J Exp Clin Cancer Res. 2018 Feb 12;37(1):24. doi: 10.1186/s13046-018-0672-z. J Exp Clin Cancer Res. 2018. PMID: 29433542 Free PMC article.
-
Breast cancer cells: Modulation by melatonin and the ubiquitin-proteasome system--a review.Mol Cell Endocrinol. 2015 Dec 5;417:1-9. doi: 10.1016/j.mce.2015.09.001. Epub 2015 Sep 9. Mol Cell Endocrinol. 2015. PMID: 26363225 Review.
Cited by
-
GREB1: An evolutionarily conserved protein with a glycosyltransferase domain links ERα glycosylation and stability to cancer.Sci Adv. 2021 Mar 17;7(12):eabe2470. doi: 10.1126/sciadv.abe2470. Print 2021 Mar. Sci Adv. 2021. PMID: 33731348 Free PMC article.
-
RBCK1 is an endogenous inhibitor for triple negative breast cancer via hippo/YAP axis.Cell Commun Signal. 2022 Oct 24;20(1):164. doi: 10.1186/s12964-022-00963-8. Cell Commun Signal. 2022. PMID: 36280829 Free PMC article.
-
Linear ubiquitination of PTEN impairs its function to promote prostate cancer progression.Oncogene. 2022 Oct;41(44):4877-4892. doi: 10.1038/s41388-022-02485-6. Epub 2022 Oct 3. Oncogene. 2022. PMID: 36192478
-
Advances in the Structural and Physiological Functions of SHARPIN.Front Immunol. 2022 Apr 25;13:858505. doi: 10.3389/fimmu.2022.858505. eCollection 2022. Front Immunol. 2022. PMID: 35547743 Free PMC article. Review.
-
Expression of the three components of linear ubiquitin assembly complex in breast cancer.PLoS One. 2018 May 15;13(5):e0197183. doi: 10.1371/journal.pone.0197183. eCollection 2018. PLoS One. 2018. PMID: 29763465 Free PMC article. Clinical Trial.
References
-
- Geschickter CF, Lewis D, Hartman CG. Tumors of the breast related to the oestrin hormone. Am J Cancer. 1934;21:828–859. - PubMed
-
- Brown RJ, Davidson NE. Adjuvant hormonal therapy for premenopausal women with breast cancer. Sem Oncol. 2006;33:657–663. - PubMed
-
- Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. - PubMed
-
- Thomas C, Gustafsson JA. The different roles of ER subtypes in cancer biology and therapy. Nat RevCancer. 2011;11:597–608. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

