Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms
- PMID: 24441097
- PMCID: PMC4057954
- DOI: 10.1038/jid.2014.28
Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms
Abstract
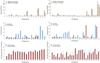
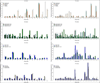
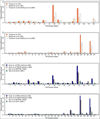
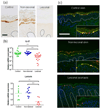
To increase our understanding of psoriasis, we used high-throughput complementary DNA sequencing (RNA-seq) to assay the transcriptomes of lesional psoriatic and normal skin. We sequenced polyadenylated RNA-derived complementary DNAs from 92 psoriatic and 82 normal punch biopsies, generating an average of ∼38 million single-end 80-bp reads per sample. Comparison of 42 samples examined by both RNA-seq and microarray revealed marked differences in sensitivity, with transcripts identified only by RNA-seq having much lower expression than those also identified by microarray. RNA-seq identified many more differentially expressed transcripts enriched in immune system processes. Weighted gene coexpression network analysis (WGCNA) revealed multiple modules of coordinately expressed epidermal differentiation genes, overlapping significantly with genes regulated by the long noncoding RNA TINCR, its target gene, staufen-1 (STAU1), the p63 target gene ZNF750, and its target KLF4. Other coordinately expressed modules were enriched for lymphoid and/or myeloid signature transcripts and genes induced by IL-17 in keratinocytes. Dermally expressed genes were significantly downregulated in psoriatic biopsies, most likely because of expansion of the epidermal compartment. These results show the power of WGCNA to elucidate gene regulatory circuits in psoriasis, and emphasize the influence of tissue architecture in both differential expression and coexpression analysis.
Conflict of interest statement
The authors declare no competing interests.
Figures
Comment in
-
RNA-seq permits a closer look at normal skin and psoriasis gene networks.J Invest Dermatol. 2014 Jul;134(7):1789-1791. doi: 10.1038/jid.2014.66. J Invest Dermatol. 2014. PMID: 24924757 Free PMC article.
References
-
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. - PubMed
-
- Bazzi H, Getz A, Mahoney MG, Ishida-Yamamoto A, Langbein L, Wahl JK, 3rd, et al. Desmoglein 4 is expressed in highly differentiated keratinocytes and trichocytes in human epidermis and hair follicle. Differentiation. 2006;74:129–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

