Temporal alterations in the secretome of the selective ligninolytic fungus Ceriporiopsis subvermispora during growth on aspen wood reveal this organism's strategy for degrading lignocellulose
- PMID: 24441164
- PMCID: PMC3993130
- DOI: 10.1128/AEM.03652-13
Temporal alterations in the secretome of the selective ligninolytic fungus Ceriporiopsis subvermispora during growth on aspen wood reveal this organism's strategy for degrading lignocellulose
Abstract
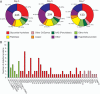
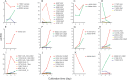
The white-rot basidiomycetes efficiently degrade all wood cell wall polymers. Generally, these fungi simultaneously degrade cellulose and lignin, but certain organisms, such as Ceriporiopsis subvermispora, selectively remove lignin in advance of cellulose degradation. However, relatively little is known about the mechanism of selective ligninolysis. To address this issue, C. subvermispora was grown in liquid medium containing ball-milled aspen, and nano-liquid chromatography-tandem mass spectrometry was used to identify and estimate extracellular protein abundance over time. Several manganese peroxidases and an aryl alcohol oxidase, both associated with lignin degradation, were identified after 3 days of incubation. A glycoside hydrolase (GH) family 51 arabinofuranosidase was also identified after 3 days but then successively decreased in later samples. Several enzymes related to cellulose and xylan degradation, such as GH10 endoxylanase, GH5_5 endoglucanase, and GH7 cellobiohydrolase, were detected after 5 days. Peptides corresponding to potential cellulose-degrading enzymes GH12, GH45, lytic polysaccharide monooxygenase, and cellobiose dehydrogenase were most abundant after 7 days. This sequential production of enzymes provides a mechanism consistent with selective ligninolysis by C. subvermispora.
Figures


References
-
- Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kues U, Kumar TK, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Duenas FJ, Sabat G, Salamov A, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. 10.1126/science.1221748 - DOI - PubMed
-
- Blanchette R. 1991. Delignification by wood-decay fungi. Annu. Rev. Phytopathol. 29:381–398. 10.1146/annurev.py.29.090191.002121 - DOI
-
- Blanchette RA, Burnes TA, Eerdmans MM, Akhtar M. 1992. Evaluating isolates of Phanerochaete chrysosporium and Ceriporiopsis subvermispora for use in biological pulping processes. Holzforschung 46:109–115. 10.1515/hfsg.1992.46.2.109 - DOI
-
- Otjen L, Blanchette R, Effland M, Leatham G. 1987. Assessment of 30 white rot basidiomycetes for selective lignin degradation. Holzforschung 41:343–349. 10.1515/hfsg.1987.41.6.343 - DOI
-
- Vanden Wymelenberg A, Gaskell J, Mozuch M, BonDurant SS, Sabat G, Ralph J, Skyba O, Mansfield SD, Blanchette RA, Grigoriev IV, Kersten PJ, Cullen D. 2011. Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species. Appl. Environ. Microbiol. 77:4499–4507. 10.1128/AEM.00508-11 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

