Autophagy is essential for cardiac morphogenesis during vertebrate development
- PMID: 24441423
- PMCID: PMC4091146
- DOI: 10.4161/auto.27649
Autophagy is essential for cardiac morphogenesis during vertebrate development
Abstract
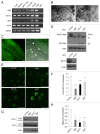
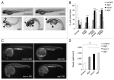
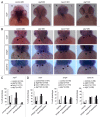
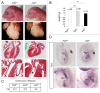
Genetic analyses indicate that autophagy, an evolutionarily conserved lysosomal degradation pathway, is essential for eukaryotic differentiation and development. However, little is known about whether autophagy contributes to morphogenesis during embryogenesis. To address this question, we examined the role of autophagy in the early development of zebrafish, a model organism for studying vertebrate tissue and organ morphogenesis. Using zebrafish that transgenically express the fluorescent autophagy reporter protein, GFP-LC3, we found that autophagy is active in multiple tissues, including the heart, during the embryonic period. Inhibition of autophagy by morpholino knockdown of essential autophagy genes (including atg5, atg7, and becn1) resulted in defects in morphogenesis, increased numbers of dead cells, abnormal heart structure, and reduced organismal survival. Further analyses of cardiac development in autophagy-deficient zebrafish revealed defects in cardiac looping, abnormal chamber morphology, aberrant valve development, and ectopic expression of critical transcription factors including foxn4, tbx5, and tbx2. Consistent with these results, Atg5-deficient mice displayed abnormal Tbx2 expression and defects in valve development and chamber septation. Thus, autophagy plays an essential, conserved role in cardiac morphogenesis during vertebrate development.
Keywords: atg5; atg7; autophagy; becn1; heart development; tbx2; zebrafish.
Figures


Similar articles
-
Expression pattern and functions of autophagy-related gene atg5 in zebrafish organogenesis.Autophagy. 2011 Dec;7(12):1514-27. doi: 10.4161/auto.7.12.18040. Autophagy. 2011. PMID: 22082871 Free PMC article.
-
Tbx5-mediated expression of Ca(2+)/calmodulin-dependent protein kinase II is necessary for zebrafish cardiac and pectoral fin morphogenesis.Dev Biol. 2009 Jun 1;330(1):175-84. doi: 10.1016/j.ydbio.2009.03.024. Epub 2009 Apr 1. Dev Biol. 2009. PMID: 19345202
-
Genetic interaction between pku300 and fbn2b controls endocardial cell proliferation and valve development in zebrafish.J Cell Sci. 2013 Mar 15;126(Pt 6):1381-91. doi: 10.1242/jcs.116996. Epub 2013 Feb 15. J Cell Sci. 2013. PMID: 23418350 Free PMC article.
-
Autophagy in the Vertebrate Inner Ear.Front Cell Dev Biol. 2017 May 26;5:56. doi: 10.3389/fcell.2017.00056. eCollection 2017. Front Cell Dev Biol. 2017. PMID: 28603711 Free PMC article. Review.
-
Uncovering the molecular and cellular mechanisms of heart development using the zebrafish.Annu Rev Genet. 2012;46:397-418. doi: 10.1146/annurev-genet-110711-155646. Epub 2012 Sep 4. Annu Rev Genet. 2012. PMID: 22974299 Free PMC article. Review.
Cited by
-
Inhibition of PRKAA/AMPK (Ser485/491) phosphorylation by crizotinib induces cardiotoxicity via perturbing autophagosome-lysosome fusion.Autophagy. 2024 Feb;20(2):416-436. doi: 10.1080/15548627.2023.2259216. Epub 2024 Jan 25. Autophagy. 2024. PMID: 37733896 Free PMC article.
-
Transgenic fluorescent zebrafish lines that have revolutionized biomedical research.Lab Anim Res. 2021 Sep 8;37(1):26. doi: 10.1186/s42826-021-00103-2. Lab Anim Res. 2021. PMID: 34496973 Free PMC article. Review.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1.Autophagy. 2021 Jan;17(1):1-382. doi: 10.1080/15548627.2020.1797280. Epub 2021 Feb 8. Autophagy. 2021. PMID: 33634751 Free PMC article.
-
Beclin-1-dependent autophagy, but not apoptosis, is critical for stem-cell-mediated endometrial programming and the establishment of pregnancy.Dev Cell. 2023 May 22;58(10):885-897.e4. doi: 10.1016/j.devcel.2023.03.013. Epub 2023 Apr 10. Dev Cell. 2023. PMID: 37040770 Free PMC article.
-
The tumor suppressor LKB1 regulates starvation-induced autophagy under systemic metabolic stress.Sci Rep. 2017 Aug 4;7(1):7327. doi: 10.1038/s41598-017-07116-9. Sci Rep. 2017. PMID: 28779098 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
