The promise and challenge of high-throughput sequencing of the antibody repertoire
- PMID: 24441474
- PMCID: PMC4113560
- DOI: 10.1038/nbt.2782
The promise and challenge of high-throughput sequencing of the antibody repertoire
Abstract
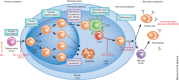
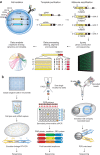
Efforts to determine the antibody repertoire encoded by B cells in the blood or lymphoid organs using high-throughput DNA sequencing technologies have been advancing at an extremely rapid pace and are transforming our understanding of humoral immune responses. Information gained from high-throughput DNA sequencing of immunoglobulin genes (Ig-seq) can be applied to detect B-cell malignancies with high sensitivity, to discover antibodies specific for antigens of interest, to guide vaccine development and to understand autoimmunity. Rapid progress in the development of experimental protocols and informatics analysis tools is helping to reduce sequencing artifacts, to achieve more precise quantification of clonal diversity and to extract the most pertinent biological information. That said, broader application of Ig-seq, especially in clinical settings, will require the development of a standardized experimental design framework that will enable the sharing and meta-analysis of sequencing data generated by different laboratories.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Janeway, C.A. Immunobiology (Garland Science, 2004).
-
- Apostoaei, A.J. & Trabalka, J.R. Review, Synthesis, and Application of Information on the Human Lymphatic System to Radiation Dosimetry for Chronic Lymphocytic Leukemia (SENES Oak Ridge, Inc., Oak Ridge, TN, 2010).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

