Insights into the binding of GABA to the insect RDL receptor from atomistic simulations: a comparison of models
- PMID: 24442887
- PMCID: PMC3927061
- DOI: 10.1007/s10822-013-9704-0
Insights into the binding of GABA to the insect RDL receptor from atomistic simulations: a comparison of models
Abstract
The resistance to dieldrin (RDL) receptor is an insect pentameric ligand-gated ion channel (pLGIC). It is activated by the neurotransmitter γ-aminobutyric acid (GABA) binding to its extracellular domain; hence elucidating the atomistic details of this interaction is important for understanding how the RDL receptor functions. As no high resolution structures are currently available, we built homology models of the extracellular domain of the RDL receptor using different templates, including the widely used acetylcholine binding protein and two pLGICs, the Erwinia Chrysanthemi ligand-gated ion channel (ELIC) and the more recently resolved GluCl. We then docked GABA into the selected three dimensional structures, which we used as starting points for classical molecular dynamics simulations. This allowed us to analyze in detail the behavior of GABA in the binding sites, including the hydrogen bond and cation-π interaction networks it formed, the conformers it visited and the possible role of water molecules in mediating the interactions; we also estimated the binding free energies. The models were all stable and showed common features, including interactions consistent with experimental data and similar to other pLGICs; differences could be attributed to the quality of the models, which increases with increasing sequence identity, and the use of a pLGIC template. We supplemented the molecular dynamics information with metadynamics, a rare event method, by exploring the free energy landscape of GABA binding to the RDL receptor. Overall, we show that the GluCl template provided the best models. GABA forming direct salt-bridges with Arg211 and Glu204, and cation-π interactions with an aromatic cage including Tyr109, Phe206 and Tyr254, represents a favorable binding arrangement, and the interaction with Glu204 can also be mediated by a water molecule.
Figures

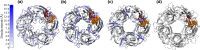


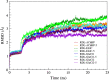

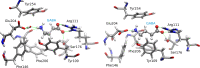
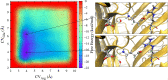
Similar articles
-
The Free Energy Landscape of GABA Binding to a Pentameric Ligand-Gated Ion Channel and Its Disruption by Mutations.J Chem Theory Comput. 2016 Jul 12;12(7):3398-406. doi: 10.1021/acs.jctc.6b00303. Epub 2016 Jun 9. J Chem Theory Comput. 2016. PMID: 27228114
-
Elucidating ligand binding and channel gating mechanisms in pentameric ligand-gated ion channels by atomistic simulations.Biochem Soc Trans. 2015 Apr;43(2):151-6. doi: 10.1042/BST20140259. Biochem Soc Trans. 2015. PMID: 25849909 Review.
-
GABA binding to an insect GABA receptor: a molecular dynamics and mutagenesis study.Biophys J. 2012 Nov 21;103(10):2071-81. doi: 10.1016/j.bpj.2012.10.016. Epub 2012 Nov 20. Biophys J. 2012. PMID: 23200041 Free PMC article.
-
Regulation of a pentameric ligand-gated ion channel by a semiconserved cationic lipid-binding site.J Biol Chem. 2021 Aug;297(2):100899. doi: 10.1016/j.jbc.2021.100899. Epub 2021 Jun 19. J Biol Chem. 2021. PMID: 34157288 Free PMC article.
-
A gating mechanism of pentameric ligand-gated ion channels.Proc Natl Acad Sci U S A. 2013 Oct 15;110(42):E3987-96. doi: 10.1073/pnas.1313785110. Epub 2013 Sep 16. Proc Natl Acad Sci U S A. 2013. PMID: 24043807 Free PMC article. Review.
Cited by
-
Transcriptome Analysis of the Central and Peripheral Nervous Systems of the Spider Cupiennius salei Reveals Multiple Putative Cys-Loop Ligand Gated Ion Channel Subunits and an Acetylcholine Binding Protein.PLoS One. 2015 Sep 14;10(9):e0138068. doi: 10.1371/journal.pone.0138068. eCollection 2015. PLoS One. 2015. PMID: 26368804 Free PMC article.
-
A Single Mutation in the Outer Lipid-Facing Helix of a Pentameric Ligand-Gated Ion Channel Affects Channel Function Through a Radially-Propagating Mechanism.Front Mol Biosci. 2021 Apr 30;8:644720. doi: 10.3389/fmolb.2021.644720. eCollection 2021. Front Mol Biosci. 2021. PMID: 33996899 Free PMC article.
-
Xenopus Oocytes: A Tool to Decipher Molecular Specificity of Insecticides towards Mammalian and Insect GABA-A Receptors.Membranes (Basel). 2022 Apr 19;12(5):440. doi: 10.3390/membranes12050440. Membranes (Basel). 2022. PMID: 35629767 Free PMC article.
-
Mutagenesis computer experiments in pentameric ligand-gated ion channels: the role of simulation tools with different resolution.Interface Focus. 2019 Jun 6;9(3):20180067. doi: 10.1098/rsfs.2018.0067. Epub 2019 Apr 19. Interface Focus. 2019. PMID: 31065340 Free PMC article. Review.
-
Insect RDL Receptor Models for Virtual Screening: Impact of the Template Conformational State in Pentameric Ligand-Gated Ion Channels.ACS Omega. 2022 Jan 5;7(2):1988-2001. doi: 10.1021/acsomega.1c05465. eCollection 2022 Jan 18. ACS Omega. 2022. PMID: 35071887 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

