Comparison of monovalent and divalent ion distributions around a DNA duplex with molecular dynamics simulation and a Poisson-Boltzmann approach
- PMID: 24443090
- PMCID: PMC4102171
- DOI: 10.1002/bip.22461
Comparison of monovalent and divalent ion distributions around a DNA duplex with molecular dynamics simulation and a Poisson-Boltzmann approach
Abstract
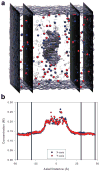
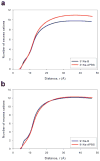
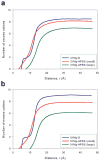
The ion atmosphere created by monovalent (Na(+) ) or divalent (Mg(2+) ) cations surrounding a B-form DNA duplex were examined using atomistic molecular dynamics (MD) simulations and the nonlinear Poisson-Boltzmann (PB) equation. The ion distributions predicted by the two methods were compared using plots of radial and two-dimensional cation concentrations and by calculating the total number of cations and net solution charge surrounding the DNA. Na(+) ion distributions near the DNA were more diffuse in PB calculations than in corresponding MD simulations, with PB calculations predicting lower concentrations near DNA groove sites and phosphate groups and a higher concentration in the region between these locations. Other than this difference, the Na(+) distributions generated by the two methods largely agreed, as both predicted similar locations of high Na(+) concentration and nearly identical values of the number of cations and the net solution charge at all distances from the DNA. In contrast, there was greater disagreement between the two methods for Mg(2+) cation concentration profiles, as both the locations and magnitudes of peaks in Mg(2+) concentration were different. Despite experimental and simulation observations that Mg(2+) typically maintains its first solvation shell when interacting with nucleic acids, modeling Mg(2+) as an unsolvated ion during PB calculations improved the agreement of the Mg(2+) ion atmosphere predicted by the two methods and allowed for values of the number of bound ions and net solution charge surrounding the DNA from PB calculations that approached the values observed in MD simulations.
Keywords: B-form DNA; Poisson-Boltzmann equation; ion atmosphere; molecular dynamics simulation; nucleic acid-cation interactions.
© 2014 Wiley Periodicals, Inc.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources