Subdiffractive microscopy: techniques, applications, and challenges
- PMID: 24443323
- PMCID: PMC3930153
- DOI: 10.1002/wsbm.1259
Subdiffractive microscopy: techniques, applications, and challenges
Abstract
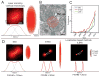
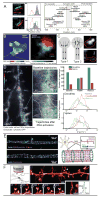
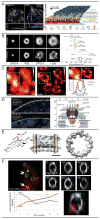
Cellular processes rely on the precise orchestration of signaling and effector molecules in space and time, yet it remains challenging to gain a comprehensive picture of the molecular organization underlying most basic biological functions. This organization often takes place at length scales below the resolving power of conventional microscopy. In recent years, several 'superresolution' fluorescence microscopic techniques have emerged that can surpass the diffraction limit of conventional microscopy by a factor of 2-20. These methods have been used to reveal previously unknown organization of macromolecular complexes and cytoskeletal structures. The resulting high-resolution view of molecular organization and dynamics is already changing our understanding of cellular processes at the systems level. However, current subdiffractive microscopic techniques are not without limitations; challenges remain to be overcome before these techniques achieve their full potential. Here, we introduce three primary types of subdiffractive microscopic techniques, consider their current limitations and challenges, and discuss recent biological applications.
© 2014 Wiley Periodicals, Inc.
Figures

Similar articles
-
Applying superresolution localization-based microscopy to neurons.Synapse. 2015 May;69(5):283-94. doi: 10.1002/syn.21806. Epub 2015 Feb 28. Synapse. 2015. PMID: 25648102 Free PMC article. Review.
-
Photoswitchable fluorescent proteins for superresolution fluorescence microscopy circumventing the diffraction limit of light.Methods Mol Biol. 2014;1076:793-812. doi: 10.1007/978-1-62703-649-8_36. Methods Mol Biol. 2014. PMID: 24108655
-
Stochastic approach to the molecular counting problem in superresolution microscopy.Proc Natl Acad Sci U S A. 2015 Jan 13;112(2):E110-8. doi: 10.1073/pnas.1408071112. Epub 2014 Dec 22. Proc Natl Acad Sci U S A. 2015. PMID: 25535361 Free PMC article.
-
Photon budget analysis for fluorescence lifetime imaging microscopy.J Biomed Opt. 2011 Aug;16(8):086007. doi: 10.1117/1.3608997. J Biomed Opt. 2011. PMID: 21895319
-
Photons in - numbers out: perspectives in quantitative fluorescence microscopy for in situ protein counting.Methods Appl Fluoresc. 2019 Jan 15;7(1):012003. doi: 10.1088/2050-6120/aaf2eb. Methods Appl Fluoresc. 2019. PMID: 30524087 Review.
Cited by
-
Applying superresolution localization-based microscopy to neurons.Synapse. 2015 May;69(5):283-94. doi: 10.1002/syn.21806. Epub 2015 Feb 28. Synapse. 2015. PMID: 25648102 Free PMC article. Review.
-
Label-free imaging of age-related cardiac structural changes in non-human primates using multiphoton nonlinear microscopy.Biomed Opt Express. 2021 Oct 19;12(11):7009-7023. doi: 10.1364/BOE.432102. eCollection 2021 Nov 1. Biomed Opt Express. 2021. PMID: 34858695 Free PMC article.
-
Tumor Retention of Enzyme-Responsive Pt(II) Drug-Loaded Nanoparticles Imaged by Nanoscale Secondary Ion Mass Spectrometry and Fluorescence Microscopy.ACS Cent Sci. 2018 Nov 28;4(11):1477-1484. doi: 10.1021/acscentsci.8b00444. Epub 2018 Oct 23. ACS Cent Sci. 2018. PMID: 30555899 Free PMC article.
-
Chromosomes at Work: Organization of Chromosome Territories in the Interphase Nucleus.J Cell Biochem. 2016 Jan;117(1):9-19. doi: 10.1002/jcb.25280. J Cell Biochem. 2016. PMID: 26192137 Free PMC article.
-
SmartScope2: Simultaneous Imaging and Reconstruction of Neuronal Morphology.Sci Rep. 2017 Aug 24;7(1):9325. doi: 10.1038/s41598-017-10067-w. Sci Rep. 2017. PMID: 28839271 Free PMC article.
References
-
- Gould TJ, Hess ST. Chapter 12: Nanoscale biological fluorescence imaging: breaking the diffraction barrier. Methods Cell Biol. 2008;89:329–358. - PubMed
-
- Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. - PubMed
-
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

