Searching for missing heritability: designing rare variant association studies
- PMID: 24443550
- PMCID: PMC3910587
- DOI: 10.1073/pnas.1322563111
Searching for missing heritability: designing rare variant association studies
Abstract
Genetic studies have revealed thousands of loci predisposing to hundreds of human diseases and traits, revealing important biological pathways and defining novel therapeutic hypotheses. However, the genes discovered to date typically explain less than half of the apparent heritability. Because efforts have largely focused on common genetic variants, one hypothesis is that much of the missing heritability is due to rare genetic variants. Studies of common variants are typically referred to as genomewide association studies, whereas studies of rare variants are often simply called sequencing studies. Because they are actually closely related, we use the terms common variant association study (CVAS) and rare variant association study (RVAS). In this paper, we outline the similarities and differences between RVAS and CVAS and describe a conceptual framework for the design of RVAS. We apply the framework to address key questions about the sample sizes needed to detect association, the relative merits of testing disruptive alleles vs. missense alleles, frequency thresholds for filtering alleles, the value of predictors of the functional impact of missense alleles, the potential utility of isolated populations, the value of gene-set analysis, and the utility of de novo mutations. The optimal design depends critically on the selection coefficient against deleterious alleles and thus varies across genes. The analysis shows that common variant and rare variant studies require similarly large sample collections. In particular, a well-powered RVAS should involve discovery sets with at least 25,000 cases, together with a substantial replication set.
Keywords: mapping disease genes; power analysis; statistical genetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
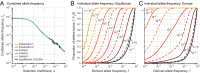

 corresponds to complete protection. (C) Relative contribution of missense vs. disruptive variants for European model. Curves for various selection coefficients show ratio of expected LOD scores for testing an excess of rare missense variants with frequency below threshold T divided by expected LOD score for disruptive variants. Values are calculated for effect size for null alleles of 1 + λ = 4. For each s, there is an optimal threshold T* at which missense alleles provide maximal relative contribution (typically ∼1.0- to 1.3-fold) (
corresponds to complete protection. (C) Relative contribution of missense vs. disruptive variants for European model. Curves for various selection coefficients show ratio of expected LOD scores for testing an excess of rare missense variants with frequency below threshold T divided by expected LOD score for disruptive variants. Values are calculated for effect size for null alleles of 1 + λ = 4. For each s, there is an optimal threshold T* at which missense alleles provide maximal relative contribution (typically ∼1.0- to 1.3-fold) (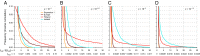
References
-
- Antonarakis SE, Chakravarti A, Cohen JC, Hardy J. Mendelian disorders and multifactorial traits: The big divide or one for all? Nat Rev Genet. 2010;11(5):380–384. - PubMed
-
- Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–165. - PubMed
-
- Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

