Molecular determinants of allosteric modulation at the M1 muscarinic acetylcholine receptor
- PMID: 24443568
- PMCID: PMC3937673
- DOI: 10.1074/jbc.M113.539080
Molecular determinants of allosteric modulation at the M1 muscarinic acetylcholine receptor
Abstract
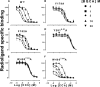
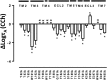
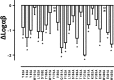
Benzylquinolone carboxylic acid (BQCA) is an unprecedented example of a selective positive allosteric modulator of acetylcholine at the M1 muscarinic acetylcholine receptor (mAChR). To probe the structural basis underlying its selectivity, we utilized site-directed mutagenesis, analytical modeling, and molecular dynamics to delineate regions of the M1 mAChR that govern modulator binding and transmission of cooperativity. We identified Tyr-85(2.64) in transmembrane domain 2 (TMII), Tyr-179 and Phe-182 in the second extracellular loop (ECL2), and Glu-397(7.32) and Trp-400(7.35) in TMVII as residues that contribute to the BQCA binding pocket at the M1 mAChR, as well as to the transmission of cooperativity with the orthosteric agonist carbachol. As such, the BQCA binding pocket partially overlaps with the previously described "common" allosteric site in the extracellular vestibule of the M1 mAChR, suggesting that its high subtype selectivity derives from either additional contacts outside this region or through a subtype-specific cooperativity mechanism. Mutation of amino acid residues that form the orthosteric binding pocket caused a loss of carbachol response that could be rescued by BQCA. Two of these residues (Leu-102(3.29) and Asp-105(3.32)) were also identified as indirect contributors to the binding affinity of the modulator. This new insight into the structural basis of binding and function of BQCA can guide the design of new allosteric ligands with tailored pharmacological properties.
Keywords: Allosteric Regulation; Drug Discovery; G Protein-coupled Receptors (GPCR); Molecular Dynamics; Muscarinic Acetylcholine Receptor; Site-directed Mutagenesis.
Figures







References
-
- Venkatakrishnan A. J., Deupi X., Lebon G., Tate C. G., Schertler G. F., Babu M. M. (2013) Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 - PubMed
-
- Lagerström M. C., Schiöth H. B. (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7, 339–357 - PubMed
-
- Hulme E. C. (2013) GPCR activation: a mutagenic spotlight on crystal structures. Trends Pharmacol. Sci. 34, 67–84 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

