Exploring accurate Poisson-Boltzmann methods for biomolecular simulations
- PMID: 24443709
- PMCID: PMC3891588
- DOI: 10.1016/j.comptc.2013.09.021
Exploring accurate Poisson-Boltzmann methods for biomolecular simulations
Abstract
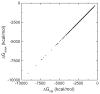
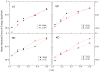
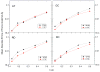
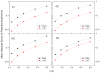
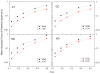
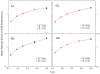
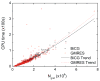
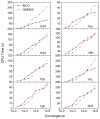
Accurate and efficient treatment of electrostatics is a crucial step in computational analyses of biomolecular structures and dynamics. In this study, we have explored a second-order finite-difference numerical method to solve the widely used Poisson-Boltzmann equation for electrostatic analyses of realistic bio-molecules. The so-called immersed interface method was first validated and found to be consistent with the classical weighted harmonic averaging method for a diversified set of test biomolecules. The numerical accuracy and convergence behaviors of the new method were next analyzed in its computation of numerical reaction field grid potentials, energies, and atomic solvation forces. Overall similar convergence behaviors were observed as those by the classical method. Interestingly, the new method was found to deliver more accurate and better-converged grid potentials than the classical method on or nearby the molecular surface, though the numerical advantage of the new method is reduced when grid potentials are extrapolated to the molecular surface. Our exploratory study indicates the need for further improving interpolation/extrapolation schemes in addition to the developments of higher-order numerical methods that have attracted most attention in the field.
Keywords: Continuum solvent models; Finite difference method; Immersed interface method; Poisson-Boltzmann equation.
Figures

Similar articles
-
Reducing grid-dependence in finite-difference Poisson-Boltzmann calculations.J Chem Theory Comput. 2012 Aug 14;8(8):2741-2751. doi: 10.1021/ct300341d. Epub 2012 Jun 18. J Chem Theory Comput. 2012. PMID: 23185142 Free PMC article.
-
On removal of charge singularity in Poisson-Boltzmann equation.J Chem Phys. 2009 Apr 14;130(14):145101. doi: 10.1063/1.3099708. J Chem Phys. 2009. PMID: 19368474
-
Highly accurate biomolecular electrostatics in continuum dielectric environments.J Comput Chem. 2008 Jan 15;29(1):87-97. doi: 10.1002/jcc.20769. J Comput Chem. 2008. PMID: 17508411
-
Polarizable atomic multipole solutes in a Poisson-Boltzmann continuum.J Chem Phys. 2007 Mar 28;126(12):124114. doi: 10.1063/1.2714528. J Chem Phys. 2007. PMID: 17411115 Free PMC article. Review.
-
Protein electrostatics: a review of the equations and methods used to model electrostatic equations in biomolecules--applications in biotechnology.Biotechnol Annu Rev. 2003;9:315-95. doi: 10.1016/s1387-2656(03)09010-0. Biotechnol Annu Rev. 2003. PMID: 14650935 Review.
Cited by
-
Robustness and Efficiency of Poisson-Boltzmann Modeling on Graphics Processing Units.J Chem Inf Model. 2019 Jan 28;59(1):409-420. doi: 10.1021/acs.jcim.8b00761. Epub 2018 Dec 31. J Chem Inf Model. 2019. PMID: 30550277 Free PMC article.
-
Numerical interpretation of molecular surface field in dielectric modeling of solvation.J Comput Chem. 2017 May 30;38(14):1057-1070. doi: 10.1002/jcc.24782. Epub 2017 Mar 20. J Comput Chem. 2017. PMID: 28318096 Free PMC article.
-
Machine-Learned Molecular Surface and Its Application to Implicit Solvent Simulations.J Chem Theory Comput. 2021 Oct 12;17(10):6214-6224. doi: 10.1021/acs.jctc.1c00492. Epub 2021 Sep 13. J Chem Theory Comput. 2021. PMID: 34516109 Free PMC article.
-
DelPhiForce, a tool for electrostatic force calculations: Applications to macromolecular binding.J Comput Chem. 2017 Apr 5;38(9):584-593. doi: 10.1002/jcc.24715. Epub 2017 Jan 28. J Comput Chem. 2017. PMID: 28130775 Free PMC article.
-
Electrostatic component of binding energy: Interpreting predictions from poisson-boltzmann equation and modeling protocols.J Comput Chem. 2016 Oct 30;37(28):2495-507. doi: 10.1002/jcc.24475. Epub 2016 Aug 21. J Comput Chem. 2016. PMID: 27546093 Free PMC article.
References
-
- Klapper I, Hagstrom R, Fine R, Sharp K, Honig B. Focusing of electric fields in the active site of copper-zinc superoxide dismutase effects of ionic strength and amino acid modification. Proteins: Struct, Funct, Genet. 1986;1:47–59. - PubMed
-
- Davis ME, McCammon JA. Solving the finite-difference linearized Poisson–Boltzmann equation – a comparison of relaxation and conjugate-gradient methods. J Comput Chem. 1989;10:386–391.
-
- Nicholls A, Honig B. A rapid finite-difference algorithm, utilizing successive over-relaxation to solve the Poisson–Boltzmann equation. J Comput Chem. 1991;12:435–445.
-
- Luty BA, Davis ME, McCammon JA. Solving the finite-difference nonlinear Poisson–Boltzmann equation. J Comput Chem. 1992;13:1114–1118.
-
- Holst M, Saied F. Multigrid solution of the Poisson–Boltzmann equation. J Comput Chem. 1993;14:105–113.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
