E2~Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis
- PMID: 24446487
- PMCID: PMC3989626
- DOI: 10.1002/embj.201386386
E2~Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis
Abstract
Pathogenic bacteria introduce effector proteins directly into the cytosol of eukaryotic cells to promote invasion and colonization. OspG, a Shigella spp. effector kinase, plays a role in this process by helping to suppress the host inflammatory response. OspG has been reported to bind host E2 ubiquitin-conjugating enzymes activated with ubiquitin (E2~Ub), a key enzyme complex in ubiquitin transfer pathways. A co-crystal structure of the OspG/UbcH5c~Ub complex reveals that complex formation has important ramifications for the activity of both OspG and the UbcH5c~Ub conjugate. OspG is a minimal kinase domain containing only essential elements required for catalysis. UbcH5c~Ub binding stabilizes an active conformation of the kinase, greatly enhancing OspG kinase activity. In contrast, interaction with OspG stabilizes an extended, less reactive form of UbcH5c~Ub. Recognizing conserved E2 features, OspG can interact with at least ten distinct human E2s~Ub. Mouse oral infection studies indicate that E2~Ub conjugates act as novel regulators of OspG effector kinase function in eukaryotic host cells.
Figures
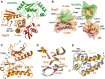
Ribbon representation of the OspG/UbcH5c-O˜Ub crystal structure determined at 2.70 Å. Ub (red) is covalently linked to UbcH5c (green) and associates in an extended conformation with OspG (orange). The OspG active site formed by a cleft between the N-and C-lobes is marked and faces away from the UbcH5c˜Ub conjugate.
‘Open book’ representation of the OspG/UbcH5c-O˜Ub interfaces. Surfaces are colored according to their interacting protein and landmark features are labeled.
Ribbon representation of the OspG kinase structure looking into the active site cleft. Secondary structure elements are labeled.
Global superposition (Supplementary Fig S1D) of OspG and PKA (PDB ID 1ATP; Zheng et al, 1993) overlay important structural and catalytic residues. OspG residues are labeled and the corresponding PKA residues are shown in Supplementary Fig S1C).
Superposition of OspG and PKA highlights differences in the arrangement of helices in the C-terminal lobe of each kinase. OspG helices are labeled. The structure of OspG is shown through the C-terminus whereas PKA has an additional 115 residues that are hidden. All PKA residue numbering used here and in the text matches PDB entry 1ATP.

2Fo-Fc composite map (blue) and Fo-Fc difference map (green) plotted at 2σ showing unknown density in the OspG active site.
Comparison with structurally aligned PKA (PDB ID 1ATP) suggests the density could be bound ATP/Mn2+.
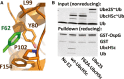
UbcH5c F62 (green) binds to a shallow pocket formed by OspG residues Y80, L99, P102, and F154.
Input: non-reducing samples of reaction mixtures prior to the addition of GST-OspG showing generation of E2˜Ub conjugates from wild-type UbcH5c, F62A-UbcH5c, and Ube2S. Pulldown: reducing SDS-PAGE shows GST-OspG only forms a high-affinity complex with wild-type UbcH5c˜Ub.
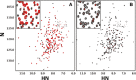
Raw heat outputs and integrated isotherms for titration of 1 mM ATP-γ-S into 50 μM GST (left), GST-OspG (center), or GST-OspG/UbcH5c-O˜Ub complex (right).
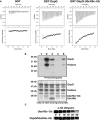
(1-5) γ32P-ATP kinase assay following phosphorylation of GST-OspG and Histone H1. 1) OspG alone, 2) premixed OspG + UbcH5c-O˜Ub, 3) premixed OspG + UbcH5c + Ub (unconjugated), 4) premixed K53M-OspG + UbcH5c-O˜Ub, 5) premixed C127R-OspG + UbcH5c-O˜Ub. The panel below depicts the proteins present in each reaction mixture.
OspG stabilizes the UbcH5c˜Ub conjugate. 5 μM samples of purified UbcH5c˜Ub or OspG/UbcH5c˜Ub were reacted with 50 mM Lysine over a 60-min time course. Non-reducing gel samples were separated by SDS-PAGE, transferred to nitrocellulose, and visualized by Western analysis for the N-terminal HA epitope on Ub. Decay of the UbcH5c˜Ub species is shown.
Comment in
-
Thinking outside the Osp(G)--kinase activation by E2-ubiquitin.EMBO J. 2014 Mar 3;33(5):403-4. doi: 10.1002/embj.201387606. Epub 2014 Jan 30. EMBO J. 2014. PMID: 24480478 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

