Microbial diversity in fecal samples depends on DNA extraction method: easyMag DNA extraction compared to QIAamp DNA stool mini kit extraction
- PMID: 24447346
- PMCID: PMC4015497
- DOI: 10.1186/1756-0500-7-50
Microbial diversity in fecal samples depends on DNA extraction method: easyMag DNA extraction compared to QIAamp DNA stool mini kit extraction
Abstract
Background: There are challenges, when extracting bacterial DNA from specimens for molecular diagnostics, since fecal samples also contain DNA from human cells and many different substances derived from food, cell residues and medication that can inhibit downstream PCR. The purpose of the study was to evaluate two different DNA extraction methods in order to choose the most efficient method for studying intestinal bacterial diversity using Denaturing Gradient Gel Electrophoresis (DGGE).
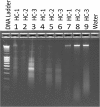
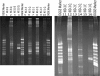
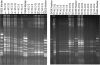
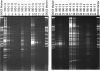
Findings: In this study, a semi-automatic DNA extraction system (easyMag®, BioMérieux, Marcy I'Etoile, France) and a manual one (QIAamp DNA Stool Mini Kit, Qiagen, Hilden, Germany) were tested on stool samples collected from 3 patients with Inflammatory Bowel disease (IBD) and 5 healthy individuals. DNA extracts obtained by the QIAamp DNA Stool Mini Kit yield a higher amount of DNA compared to DNA extracts obtained by easyMag® from the same fecal samples. Furthermore, DNA extracts obtained using easyMag® seemed to contain inhibitory compounds, since in order to perform a successful PCR-analysis, the sample should be diluted at least 10 times. DGGE performed on PCR from DNA extracted by QIAamp DNA Stool Mini Kit DNA was very successful.
Conclusion: QIAamp DNA Stool Mini Kit DNA extracts are optimal for DGGE runs and this extraction method yields a higher amount of DNA compared to easyMag®.
Figures

Similar articles
-
Evaluation of QIAamp DNA Stool Mini Kit for ecological studies of gut microbiota.J Microbiol Methods. 2003 Jul;54(1):13-20. doi: 10.1016/s0167-7012(02)00260-9. J Microbiol Methods. 2003. PMID: 12732417
-
Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens.Nutr J. 2010 May 22;9:23. doi: 10.1186/1475-2891-9-23. Nutr J. 2010. PMID: 20492702 Free PMC article.
-
A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples.J Microbiol Methods. 2013 Aug;94(2):103-110. doi: 10.1016/j.mimet.2013.05.008. Epub 2013 May 14. J Microbiol Methods. 2013. PMID: 23684993 Free PMC article.
-
Evaluation of fecal DNA extraction protocols for human gut microbiome studies.BMC Microbiol. 2020 Jul 17;20(1):212. doi: 10.1186/s12866-020-01894-5. BMC Microbiol. 2020. PMID: 32680572 Free PMC article.
-
Microbial diversity in the human intestine and novel insights from metagenomics.Front Biosci (Landmark Ed). 2009 Jan 1;14(9):3214-21. doi: 10.2741/3445. Front Biosci (Landmark Ed). 2009. PMID: 19273267 Review.
Cited by
-
Inhibition of Tumor Growth by Dietary Indole-3-Carbinol in a Prostate Cancer Xenograft Model May Be Associated with Disrupted Gut Microbial Interactions.Nutrients. 2019 Feb 22;11(2):467. doi: 10.3390/nu11020467. Nutrients. 2019. PMID: 30813350 Free PMC article.
-
Alteration of gut microbiota by a Westernized lifestyle and its correlation with insulin resistance in non-diabetic Japanese men.J Diabetes Investig. 2019 Nov;10(6):1463-1470. doi: 10.1111/jdi.13048. Epub 2019 Apr 19. J Diabetes Investig. 2019. PMID: 30901505 Free PMC article.
-
A randomized trial to evaluate the impact of copra meal hydrolysate on gastrointestinal symptoms and gut microbiome.PeerJ. 2021 Sep 15;9:e12158. doi: 10.7717/peerj.12158. eCollection 2021. PeerJ. 2021. PMID: 34616618 Free PMC article.
-
Identification of DNA methylation markers for early detection of CRC indicates a role for nervous system-related genes in CRC.Clin Epigenetics. 2021 Apr 15;13(1):80. doi: 10.1186/s13148-021-01067-9. Clin Epigenetics. 2021. PMID: 33858496 Free PMC article.
-
Molecular Insights Into the Role of Gut Microbiota in Antibiotic Therapy Selection and Resistance Mitigation.Cureus. 2023 Dec 11;15(12):e50318. doi: 10.7759/cureus.50318. eCollection 2023 Dec. Cureus. 2023. PMID: 38089944 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

