Duplex formation between the sRNA DsrA and rpoS mRNA is not sufficient for efficient RpoS synthesis at low temperature
- PMID: 24448230
- PMCID: PMC3917986
- DOI: 10.4161/rna.27100
Duplex formation between the sRNA DsrA and rpoS mRNA is not sufficient for efficient RpoS synthesis at low temperature
Abstract
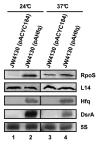
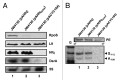
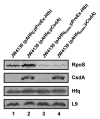
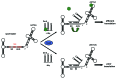
At low temperatures the Escherichia coli rpoS mRNA, encoding the stationary phase sigma factor RpoS, forms an intramolecular secondary structure (iss) that impedes translation initiation. Under these conditions the small RNA DsrA, which is stabilzed by Hfq, forms a duplex with rpoS mRNA sequences opposite of the ribosome-binding site (rbs). Both the DEAD box helicase CsdA and Hfq have been implicated in DsrA·rpoS duplex formation. Hfq binding to A-rich sequences in the rpoS leader has been suggested to restructure the mRNA, and thereby to accelerate DsrA·rpoS duplex formation, which, in turn, was deemed to free the rpoS rbs and to permit ribosome loading on the mRNA. Several experiments designed to elucidate the role of Hfq in DsrA-mediated translational activation of rpoS mRNA have been conducted in vitro. Here, we assessed RpoS synthesis in vivo to further study the role of Hfq in rpoS regulation. We show that RpoS synthesis was reduced when DsrA was ectopically overexpressed at 24 °C in the absence of Hfq despite of DsrA·rpoS duplex formation. This observation indicated that DsrA·rpoS annealing may not be sufficient for efficient ribosome loading on rpoS mRNA. In addition, a HfqG29A mutant protein was employed, which is deficient in binding to A-rich sequences present in the rpoS leader but proficient in DsrA binding. We show that DsrA·rpoS duplex formation occurs in the presence of the HfqG29A mutant protein at low temperature, whereas synthesis of RpoS was greatly diminished. RNase T1 footprinting studies of DsrA·rpoS duplexes in the absence and presence of Hfq or HfqG29A indicated that Hfq is required to resolve a stem-loop structure in the immediate coding region of rpoS mRNA. These in vivo studies corroborate the importance of the A-rich sequences in the rpoS leader and strongly suggest that Hfq, besides stabilizing DsrA and accelerating DsrA·rpoS duplex formation, is also required to convert the rpoS mRNA into a translationally competent form.
Keywords: DsrA; Hfq; riboregulation; rpoS; translational activation.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
