Label-free mass spectrometry exploits dozens of detected peptides to quantify lamins in wildtype and knockdown cells
- PMID: 24448480
- PMCID: PMC3925690
- DOI: 10.4161/nucl.27413
Label-free mass spectrometry exploits dozens of detected peptides to quantify lamins in wildtype and knockdown cells
Abstract
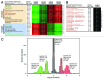
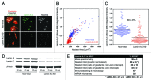
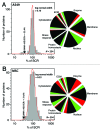
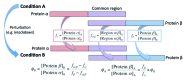
Label-free quantitation and characterization of proteins by mass spectrometry (MS) is now feasible, especially for moderately expressed structural proteins such as lamins that typically yield dozens of tryptic peptides from tissue cells. Using standard cell culture samples, we describe general algorithms for quantitative analysis of peptides identified in liquid chromatography tandem mass spectrometry (LC-MS/MS). The algorithms were foundational to the discovery that the absolute stoichiometry of A-type to B-type lamins scales with tissue stiffness (Swift et al., Science 2013). Isoform dominance helps make sense of why mutations and changes with age of mechanosensitive lamin-A,C only affect "stiff" tissues such as heart, muscle, bone, or even fat, but not brain. A Peak Ratio Fingerprinting (PRF) algorithm is elaborated here through its application to lamin-A,C knockdown. After demonstrating the large dynamic range of PRF using calibrated mixtures of human and mouse lysates, we validate measurements of partial knockdown with standard cell biology analyses using quantitative immunofluorescence and immunoblotting. Optimal sets of MS-detected peptides as determined by PRF demonstrate that the strongest peptide signals are not necessarily the most reliable for quantitation. After lamin-A,C knockdown, PRF computes an invariant set of "housekeeping" proteins as part of a broader proteomic analysis that also shows the proteome of mesenchymal stem cells (MSCs) is more broadly perturbed than that of a human epithelial cancer line (A549s), with particular variation in nuclear and cytoskeletal proteins. These methods offer exciting prospects for basic and clinical studies of lamin-A,C as well as other MS-detectable proteins.
Keywords: isoform; label-free; lamin; mass spectrometry; normalization; proteomics; spliceoform.
Figures

Comment on
-
Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation.Science. 2013 Aug 30;341(6149):1240104. doi: 10.1126/science.1240104. Science. 2013. PMID: 23990565 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
