Fragile X syndrome due to a missense mutation
- PMID: 24448548
- PMCID: PMC4169535
- DOI: 10.1038/ejhg.2013.311
Fragile X syndrome due to a missense mutation
Abstract
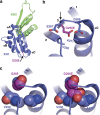
Fragile X syndrome is a common inherited form of intellectual disability and autism spectrum disorder. Most patients exhibit a massive CGG-repeat expansion mutation in the FMR1 gene that silences the locus. In over two decades since the discovery of FMR1, only a single missense mutation (p.(Ile304Asn)) has been reported as causing fragile X syndrome. Here we describe a 16-year-old male presenting with fragile X syndrome but without the repeat expansion mutation. Rather, we find a missense mutation, c.797G>A, that replaces glycine 266 with glutamic acid (p.(Gly266Glu)). The Gly266Glu FMR protein abolished many functional properties of the protein. This patient highlights the diagnostic utility of FMR1 sequencing.
Figures

References
-
- Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Ann Rev Pathol. 2012;7:219–245. - PubMed
-
- Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. - PubMed
-
- Fu YH, Kuhl DP, Pizzuti A, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

