Enhanced enzyme kinetic stability by increasing rigidity within the active site
- PMID: 24448805
- PMCID: PMC3953309
- DOI: 10.1074/jbc.M113.536045
Enhanced enzyme kinetic stability by increasing rigidity within the active site
Abstract
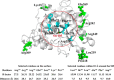
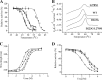
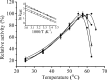
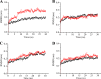
Enzyme stability is an important issue for protein engineers. Understanding how rigidity in the active site affects protein kinetic stability will provide new insight into enzyme stabilization. In this study, we demonstrated enhanced kinetic stability of Candida antarctica lipase B (CalB) by mutating the structurally flexible residues within the active site. Six residues within 10 Å of the catalytic Ser(105) residue with a high B factor were selected for iterative saturation mutagenesis. After screening 2200 colonies, we obtained the D223G/L278M mutant, which exhibited a 13-fold increase in half-life at 48 °C and a 12 °C higher T50(15), the temperature at which enzyme activity is reduced to 50% after a 15-min heat treatment. Further characterization showed that global unfolding resistance against both thermal and chemical denaturation also improved. Analysis of the crystal structures of wild-type CalB and the D223G/L278M mutant revealed that the latter formed an extra main chain hydrogen bond network with seven structurally coupled residues within the flexible α10 helix that are primarily involved in forming the active site. Further investigation of the relative B factor profile and molecular dynamics simulation confirmed that the enhanced rigidity decreased fluctuation of the active site residues at high temperature. These results indicate that enhancing the rigidity of the flexible segment within the active site may provide an efficient method for improving enzyme kinetic stability.
Keywords: Active Site; Crystal Structure; Lipase; Local Rigidity; Mutagenesis; Protein Design; Protein Stability.
Figures


References
-
- Yi Z. L., Pei X. Q., Wu Z. L. (2011) Introduction of glycine and proline residues onto protein surface increases the thermostability of endoglucanase CelA from Clostridium thermocellum. Bioresour. Technol. 102, 3636–3638 - PubMed
-
- Liang X., Bian Y., Tang X. F., Xiao G., Tang B. (2010) Enhancement of keratinolytic activity of a thermophilic subtilase by improving its autolysis resistance and thermostability under reducing conditions. Appl. Microbiol. Biotechnol. 87, 999–1006 - PubMed
-
- Yang G., Bai A., Gao L., Zhang Z., Zheng B., Feng Y. (2009) Glu88 in the non-catalytic domain of acylpeptide hydrolase plays dual roles: charge neutralization for enzymatic activity and formation of salt bridge for thermodynamic stability. Biochim. Biophys. Acta 1794, 94–102 - PubMed
-
- Voutilainen S. P., Boer H., Alapuranen M., Jänis J., Vehmaanperä J., Koivula A. (2009) Improving the thermostability and activity of Melanocarpus albomyces cellobiohydrolase Cel7B. Appl. Microbiol. Biotechnol. 83, 261–272 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

