CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner
- PMID: 24449877
- PMCID: PMC3918772
- DOI: 10.1073/pnas.1319953111
CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner
Abstract
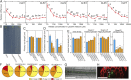
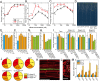
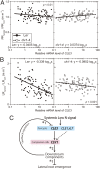
Morphological plasticity of root systems is critically important for plant survival because it allows plants to optimize their capacity to take up water and nutrients from the soil environment. Here we show that a signaling module composed of nitrogen (N)-responsive CLE (CLAVATA3/ESR-related) peptides and the CLAVATA1 (CLV1) leucine-rich repeat receptor-like kinase is expressed in the root vasculature in Arabidopsis thaliana and plays a crucial role in regulating the expansion of the root system under N-deficient conditions. CLE1, -3, -4, and -7 were induced by N deficiency in roots, predominantly expressed in root pericycle cells, and their overexpression repressed the growth of lateral root primordia and their emergence from the primary root. In contrast, clv1 mutants showed progressive outgrowth of lateral root primordia into lateral roots under N-deficient conditions. The clv1 phenotype was reverted by introducing a CLV1 promoter-driven CLV1:GFP construct producing CLV1:GFP fusion proteins in phloem companion cells of roots. The overaccumulation of CLE2, -3, -4, and -7 in clv1 mutants suggested the amplitude of the CLE peptide signals being feedback-regulated by CLV1. When CLE3 was overexpressed under its own promoter in wild-type plants, the length of lateral roots was negatively correlated with increasing CLE3 mRNA levels; however, this inhibitory action of CLE3 was abrogated in the clv1 mutant background. Our findings identify the N-responsive CLE-CLV1 signaling module as an essential mechanism restrictively controlling the expansion of the lateral root system in N-deficient environments.
Keywords: nitrogen signaling; root morphology; root system architecture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol. 2003;6(3):280–287. - PubMed
-
- Hodge A. The plastic plant: Root responses to heterogeneous supplies of nutrients. New Phytol. 2004;162(1):9–24.
-
- Robinson D. The responses of plants to non-uniform supplies of nutrients. New Phytol. 1994;127(4):635–674. - PubMed
-
- Drew MC. Comparison of the effects of a localised supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot, in barley. New Phytol. 1975;75(3):479–490.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

