Induced multipotency in adult keratinocytes through down-regulation of ΔNp63 or DGCR8
- PMID: 24449888
- PMCID: PMC3918754
- DOI: 10.1073/pnas.1319743111
Induced multipotency in adult keratinocytes through down-regulation of ΔNp63 or DGCR8
Abstract
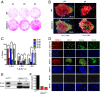
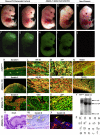
The roles of microRNAs (miRNAs) and the miRNA processing machinery in the regulation of stem cell biology are not well understood. Here, we show that the p53 family member and p63 isoform, ΔNp63, is a transcriptional activator of a cofactor critical for miRNA processing (DGCR8). This regulation gives rise to a unique miRNA signature resulting in reprogramming cells to multipotency. Strikingly, ΔNp63(-/-) epidermal cells display profound defects in terminal differentiation and express a subset of markers and miRNAs present in embryonic stem cells and fibroblasts induced to pluripotency using Yamanaka factors. Moreover, ΔNp63(-/-) epidermal cells transduced with an inducible DGCR8 plasmid can differentiate into multiple cell fates in vitro and in vivo. We found that human primary keratinocytes depleted of ΔNp63 or DGCR8 can be reprogrammed in 6 d and express a unique miRNA and gene expression signature that is similar but not identical to human induced pluripotent stem cells. Our data reveal a role for ΔNp63 in the transcriptional regulation of DGCR8 to reprogram adult somatic cells into multipotent stem cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

