Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases
- PMID: 24449932
- PMCID: PMC3596956
- DOI: 10.1021/bi301600z
Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases
Erratum in
- Biochemistry. 2013 Jun 11;52(23):4105
Abstract
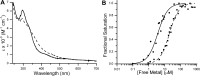
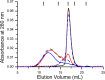
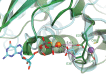
GTPases are critical molecular switches involved in a wide range of biological functions. Recent phylogenetic and genomic analyses of the large, mostly uncharacterized COG0523 subfamily of GTPases revealed a link between some COG0523 proteins and metal homeostasis pathways. In this report, we detail the bioinorganic characterization of YjiA, a representative member of COG0523 subgroup 9 and the only COG0523 protein to date with high-resolution structural information. We find that YjiA is capable of binding several types of transition metals with dissociation constants in the low micromolar range and that metal binding affects both the oligomeric structure and GTPase activity of the enzyme. Using a combination of X-ray crystallography and site-directed mutagenesis, we identify, among others, a metal-binding site adjacent to the nucleotide-binding site in the GTPase domain that involves a conserved cysteine and several glutamate residues. Mutations of the coordinating residues decrease the impact of metal, suggesting that metal binding to this site is responsible for modulating the GTPase activity of the protein. These findings point toward a regulatory function for these COG0523 GTPases that is responsive to their metal-bound state.
Figures



Similar articles
-
YeiR: a metal-binding GTPase from Escherichia coli involved in metal homeostasis.Metallomics. 2012 May;4(5):488-97. doi: 10.1039/c2mt20012k. Epub 2012 Apr 17. Metallomics. 2012. PMID: 22511334 Free PMC article.
-
COG0523 proteins: a functionally diverse family of transition metal-regulated G3E P-loop GTP hydrolases from bacteria to man.Metallomics. 2021 Aug 13;13(8):mfab046. doi: 10.1093/mtomcs/mfab046. Metallomics. 2021. PMID: 34302342 Free PMC article.
-
The iron-type nitrile hydratase activator protein is a GTPase.Biochem J. 2017 Jan 15;474(2):247-258. doi: 10.1042/BCJ20160884. Epub 2016 Nov 2. Biochem J. 2017. PMID: 27807009
-
Structural insights into the GTPase domain of Escherichia coli MnmE protein.Proteins. 2007 Feb 15;66(3):726-39. doi: 10.1002/prot.21186. Proteins. 2007. PMID: 17143896
-
Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function.Proteins. 2004 Feb 1;54(2):371-4. doi: 10.1002/prot.10430. Proteins. 2004. PMID: 14696199 No abstract available.
Cited by
-
Moving metals: How microbes deliver metal cofactors to metalloproteins.Mol Microbiol. 2023 Oct;120(4):547-554. doi: 10.1111/mmi.15117. Epub 2023 Jul 5. Mol Microbiol. 2023. PMID: 37408317 Free PMC article.
-
Zur: Zinc-Sensing Transcriptional Regulator in a Diverse Set of Bacterial Species.Pathogens. 2021 Mar 15;10(3):344. doi: 10.3390/pathogens10030344. Pathogens. 2021. PMID: 33804265 Free PMC article. Review.
-
Relationship between Ni(II) and Zn(II) coordination and nucleotide binding by the Helicobacter pylori [NiFe]-hydrogenase and urease maturation factor HypB.J Biol Chem. 2014 Feb 14;289(7):3828-41. doi: 10.1074/jbc.M113.502781. Epub 2013 Dec 12. J Biol Chem. 2014. PMID: 24338018 Free PMC article.
-
Interplay between the Zur Regulon Components and Metal Resistance in Cupriavidus metallidurans.J Bacteriol. 2019 Jul 10;201(15):e00192-19. doi: 10.1128/JB.00192-19. Print 2019 Aug 1. J Bacteriol. 2019. PMID: 31109989 Free PMC article.
-
Crystal structures of AztD provide mechanistic insights into direct zinc transfer between proteins.Commun Biol. 2019 Aug 9;2:308. doi: 10.1038/s42003-019-0542-z. eCollection 2019. Commun Biol. 2019. PMID: 31428696 Free PMC article.
References
-
- Clementi N.; Polacek N. (2010) Ribosome-associated GTPases: The role of RNA for GTPase activation. RNA Biol. 7, 521–527. - PubMed
-
- Bourne H. R.; Sanders D. A.; McCormick F. (1990) The GTPase superfamily: A conserved switch for diverse cell functions. Nature 348, 125–132. - PubMed
-
- Leipe D. D.; Wolf Y. I.; Koonin E. V.; Aravind L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72. - PubMed
-
- Saraste M.; Sibbald P. R.; Wittinghofer A. (1990) The P-loop: A common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

