Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation
- PMID: 24450871
- PMCID: PMC3971609
- DOI: 10.1186/2049-2618-1-19
Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation
Erratum in
- Microbiome. 2014;2:21. Gao, Zhan [added]
Abstract
Background: The lung microbiome of healthy individuals frequently harbors oral organisms. Despite evidence that microaspiration is commonly associated with smoking-related lung diseases, the effects of lung microbiome enrichment with upper airway taxa on inflammation has not been studied. We hypothesize that the presence of oral microorganisms in the lung microbiome is associated with enhanced pulmonary inflammation. To test this, we sampled bronchoalveolar lavage (BAL) from the lower airways of 29 asymptomatic subjects (nine never-smokers, 14 former-smokers, and six current-smokers). We quantified, amplified, and sequenced 16S rRNA genes from BAL samples by qPCR and 454 sequencing. Pulmonary inflammation was assessed by exhaled nitric oxide (eNO), BAL lymphocytes, and neutrophils.
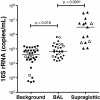
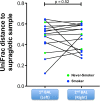
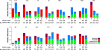
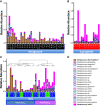
Results: BAL had lower total 16S than supraglottic samples and higher than saline background. Bacterial communities in the lower airway clustered in two distinct groups that we designated as pneumotypes. The rRNA gene concentration and microbial community of the first pneumotype was similar to that of the saline background. The second pneumotype had higher rRNA gene concentration and higher relative abundance of supraglottic-characteristic taxa (SCT), such as Veillonella and Prevotella, and we called it pneumotypeSCT. Smoking had no effect on pneumotype allocation, α, or β diversity. PneumotypeSCT was associated with higher BAL lymphocyte-count (P= 0.007), BAL neutrophil-count (P= 0.034), and eNO (P= 0.022).
Conclusion: A pneumotype with high relative abundance of supraglottic-characteristic taxa is associated with enhanced subclinical lung inflammation.
Figures


References
-
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;1:1578–1593. doi: 10.1016/j.cell.2012.04.037. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

