Genetic incorporation of seven ortho-substituted phenylalanine derivatives
- PMID: 24451054
- PMCID: PMC3997995
- DOI: 10.1021/cb400917a
Genetic incorporation of seven ortho-substituted phenylalanine derivatives
Abstract
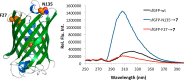
Seven phenylalanine derivatives with small ortho substitutions were genetically encoded in Escherichia coli and mammalian cells at an amber codon using a previously reported, rationally designed pyrrolysyl-tRNA synthetase mutant (PylRS(N346A/C348A)) coupled with tRNACUAPyl. Ortho substitutions of the phenylalanine derivatives reported herein include three halides, methyl, methoxy, nitro, and nitrile. These compounds have the potential for use in multiple biochemical and biophysical applications. Specifically, we demonstrated that o-cyano-phenylalanine could be used as a selective sensor to probe the local environment of proteins and applied this to study protein folding/unfolding. For six of these compounds this constitutes the first report of their genetic incorporation in living cells. With these compounds the total number of substrates available for PylRS(N346A/C348A) is increased to nearly 40, which demonstrates that PylRS(N346A/C348A) is able to recognize phenylalanine with a substitution at any side-chain aromatic position as a substrate. To our knowledge, PylRS(N346A/C348A) is the only aminoacyl-tRNA synthetase with such a high substrate promiscuity.
Figures




Similar articles
-
Genetic incorporation of twelve meta-substituted phenylalanine derivatives using a single pyrrolysyl-tRNA synthetase mutant.ACS Chem Biol. 2013 Feb 15;8(2):405-15. doi: 10.1021/cb300512r. Epub 2012 Nov 19. ACS Chem Biol. 2013. PMID: 23138887 Free PMC article.
-
Site-specific incorporation of unnatural amino acids into urate oxidase in Escherichia coli.Protein Sci. 2008 Oct;17(10):1827-33. doi: 10.1110/ps.034587.108. Epub 2008 Jul 2. Protein Sci. 2008. PMID: 18596202 Free PMC article.
-
The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives.Mol Biosyst. 2011 Mar;7(3):714-7. doi: 10.1039/c0mb00217h. Epub 2011 Jan 14. Mol Biosyst. 2011. PMID: 21234492
-
Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli.J Am Chem Soc. 2002 Aug 7;124(31):9026-7. doi: 10.1021/ja027007w. J Am Chem Soc. 2002. PMID: 12148987
-
An expanding genetic code.Trends Genet. 2004 Dec;20(12):625-30. doi: 10.1016/j.tig.2004.09.013. Trends Genet. 2004. PMID: 15522458 Review.
Cited by
-
Facile Removal of Leader Peptides from Lanthipeptides by Incorporation of a Hydroxy Acid.J Am Chem Soc. 2015 Jun 10;137(22):6975-8. doi: 10.1021/jacs.5b04681. Epub 2015 Jun 1. J Am Chem Soc. 2015. PMID: 26006047 Free PMC article.
-
Genetic Encoding of Three Distinct Noncanonical Amino Acids Using Reprogrammed Initiator and Nonsense Codons.ACS Chem Biol. 2021 Apr 16;16(4):766-774. doi: 10.1021/acschembio.1c00120. Epub 2021 Mar 16. ACS Chem Biol. 2021. PMID: 33723984 Free PMC article.
-
A Designed, Highly Efficient Pyrrolysyl-tRNA Synthetase Mutant Binds o-Chlorophenylalanine Using Two Halogen Bonds.J Mol Biol. 2022 Apr 30;434(8):167534. doi: 10.1016/j.jmb.2022.167534. Epub 2022 Mar 9. J Mol Biol. 2022. PMID: 35278475 Free PMC article.
-
Engineering of enzymes using non-natural amino acids.Biosci Rep. 2022 Aug 31;42(8):BSR20220168. doi: 10.1042/BSR20220168. Biosci Rep. 2022. PMID: 35856922 Free PMC article. Review.
-
Using genetically incorporated unnatural amino acids to control protein functions in mammalian cells.Essays Biochem. 2019 Jul 3;63(2):237-266. doi: 10.1042/EBC20180042. Print 2019 Jul 3. Essays Biochem. 2019. PMID: 31092687 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

