An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome
- PMID: 24451062
- PMCID: PMC3971625
- DOI: 10.1186/2049-2618-1-25
An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome
Abstract
Background: Our knowledge of microbial diversity in the human oral cavity has vastly expanded during the last two decades of research. However, much of what is known about the behavior of oral species to date derives from pure culture approaches and the studies combining several cultivated species, which likely does not fully reflect their function in complex microbial communities. It has been shown in studies with a limited number of cultivated species that early oral biofilm development occurs in a successional manner and that continuous low pH can lead to an enrichment of aciduric species. Observations that in vitro grown plaque biofilm microcosms can maintain similar pH profiles in response to carbohydrate addition as plaque in vivo suggests a complex microbial community can be established in the laboratory. In light of this, our primary goal was to develop a robust in vitro biofilm-model system from a pooled saliva inoculum in order to study the stability, reproducibility, and development of the oral microbiome, and its dynamic response to environmental changes from the community to the molecular level.
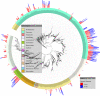
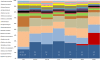
Results: Comparative metagenomic analyses confirmed a high similarity of metabolic potential in biofilms to recently available oral metagenomes from healthy subjects as part of the Human Microbiome Project. A time-series metagenomic analysis of the taxonomic community composition in biofilms revealed that the proportions of major species at 3 hours of growth are maintained during 48 hours of biofilm development. By employing deep pyrosequencing of the 16S rRNA gene to investigate this biofilm model with regards to bacterial taxonomic diversity, we show a high reproducibility of the taxonomic carriage and proportions between: 1) individual biofilm samples; 2) biofilm batches grown at different dates; 3) DNA extraction techniques and 4) research laboratories.
Conclusions: Our study demonstrates that we now have the capability to grow stable oral microbial in vitro biofilms containing more than one hundred operational taxonomic units (OTU) which represent 60-80% of the original inoculum OTU richness. Previously uncultivated Human Oral Taxa (HOT) were identified in the biofilms and contributed to approximately one-third of the totally captured 16S rRNA gene diversity. To our knowledge, this represents the highest oral bacterial diversity reported for an in vitro model system so far. This robust model will help investigate currently uncultivated species and the known virulence properties for many oral pathogens not solely restricted to pure culture systems, but within multi-species biofilms.
Figures



References
-
- Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr Issues Mol Biol. 2005;1:95–107. - PubMed
-
- Stephan RM, Hemmens ES. Studies of changes in pH produced by pure cultures of oral micro-organisms; effects of varying the microbic cell concentration; comparison of different micro-organisms and different substrates; some effects of mixing certain micro-organisms. J Dent Res. 1947;1:15–41. doi: 10.1177/00220345470260010201. - DOI - PubMed
-
- Stephan RM, Hemmens E. pH studies on oral micro-organisms. J Dent Res. 1946;1:172. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

